Joint spatial and spatiotemporal methods for modeling infectious diseases: a systematic review
Lameck Ondieki Agasa, Leyla Abdullahi, Samuel Mongare, Thomas Achia, Wycliffe Kipkoech Cheruiyot, Antony Karanja
Corresponding author: Lameck Agasa, Department of Global and Public Health, Faculty of Health Sciences, University of Nairobi, Nairobi, Kenya 
Received: 06 Nov 2023 - Accepted: 31 Mar 2024 - Published: 25 Jul 2024
Domain: Biostatistics,Epidemiology,Infectious diseases epidemiology
Keywords: Spatial and temporal modeling, joint disease models, covariates, Bayesian paradigm, disease surveillance
©Lameck Ondieki Agasa et al. PAMJ-One Health (ISSN: 2707-2800). This is an Open Access article distributed under the terms of the Creative Commons Attribution International 4.0 License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cite this article: Lameck Ondieki Agasa et al. Joint spatial and spatiotemporal methods for modeling infectious diseases: a systematic review. PAMJ-One Health. 2024;14:14. [doi: 10.11604/pamj-oh.2024.14.14.42127]
Available online at: https://www.one-health.panafrican-med-journal.com/content/article/14/14/full
Review 
Joint spatial and spatiotemporal methods for modeling infectious diseases: a systematic review
Joint spatial and spatiotemporal methods for modelling infectious diseases: a systematic review
![]() Lameck Ondieki Agasa1,2,&, Leyla Abdullahi3,
Lameck Ondieki Agasa1,2,&, Leyla Abdullahi3, ![]() Samuel Mongare1, Thomas Achia4, Wycliffe Kipkoech Cheruiyot5, Antony Karanja6
Samuel Mongare1, Thomas Achia4, Wycliffe Kipkoech Cheruiyot5, Antony Karanja6
&Corresponding author
Introduction: infectious diseases present significant global public health challenges, sharing common transmission cycles, clinical manifestations, and epidemiological characteristics. Understanding these diseases collectively can offer valuable insights into their dynamics. This study aims to synthesize methodologies used in spatial and temporal modeling of diseases, focusing on identifying common covariates and factors influencing joint disease modeling.
Methods: a systematic search was conducted in June 2023 across electronic databases (Scopus, PubMed, Web of Science, Cochrane Library) using specified terms. Articles published in English from January 2000 to April 2023 were included. Screening, data extraction, and critical appraisal were independently performed by two reviewers. The review employed a modified quality assessment tool with a median score of 10/12 for the included studies.
Results: seventeen articles met inclusion criteria, with Bayesian methods prominently utilized in 10 studies. These studies employed generalized linear mixed models integrating spatial random effects to jointly model diseases. Nine studies conducted simulations to validate their findings. Environmental and climatic variables were frequently identified as significant covariates in these models.
Conclusion: joint modeling approaches incorporating Bayesian frameworks and spatial random effects offer robust methodologies for disease surveillance and understanding transmission dynamics across time and space. Further advancements in methodological approaches will be essential for enhancing disease modeling accuracy and informing effective public health interventions.
Spatiotemporal modeling and analysis aim to contribute to our understanding of disease transmission dynamics, prediction of epidemic courses, assessment of control measures, and determination of epidemic sources in both space and time, as well as their interaction [1-3]. However, there has been limited progress in spatial dynamics modeling of infectious diseases [4,5]. Most proposed approaches employ population and individual level methods to design mathematical models, statistical models, and spatial simulation models [6]. The Susceptible-Infection-Recovery (SIR) model, introduced by Kermack and McKendrick in 1927, is a commonly used model for epidemic disease transmission, by Kreck et al. [7]. Modeling diseases jointly allows researchers to identify potential risk factors associated with different diseases, providing more robust and convincing evidence for the underlying risks in each disease under study [8,9]. Additionally, it enables the understanding of interactions among two or more diseases [10]. Aswi et al. [11] emphasized the importance of shared components among diseases when mapping multiple diseases. Recent advancements have strengthened the inference of disease data by extending spatial models to include aspects like time, space, and space-time interactions. For example, Costa et al. [12] modeled dengue fever, Zika, and chikungunya in Brazil, highlighting the significance of spatial analysis in determining priority areas for public health interventions [13]. However, the joint transmission of emerging diseases remains understudied, leading to knowledge gaps.
Numerous spatial models have been proposed, ranging from univariate data-based models to spatial multivariate models [14]. Bayesian spatial and spatiotemporal modeling approaches were systematically reviewed by Aswi et al. [11], revealing their ability to incorporate a wider range of variance components at different levels in the model, facilitating a comprehensive assessment of prediction uncertainty. Several Bayesian spatiotemporal models have been applied to study joint diseases, including the Generalized Linear Mixed Model (GLMM) with both spatial and temporal components proposed by Wah et al. [15] and the Generalized Linear Mixed Model (GLMM) with only spatial random effects studied by Knorr-Held et al. [16]. Efforts have been made to model two diseases with a shared component [17], extending the single-disease modeling approaches studied by Knorr-Held et al. [16]. The shared component model breaks down the two diseases into three spatial components that allow for spatial random effects: one shared by both diseases and two disease-specific components reflecting residual spatial variation unique to each disease [18]. This extends the work of Wakefield et al. [19] by proposing an alternative formulation using a bivariate Bayesian Poisson mixed model to jointly model two diseases, addressing the assumptions from [20,21] of shared and disease-specific spatial components being assumed to be independent and ignoring the possibility of interactions between underlying covariates with significant spatial structure.
Spatial, temporal, and statistical methodologies in health play a significant role in determining the spread and transmission of diseases, particularly in monitoring and surveillance efforts [15,20]. Advances in joint spatiotemporal modeling of infectious diseases have been driven by improvements in geostatistical techniques [22]. Despite this progress, there have been no prior reviews on the methodological modeling of joint diseases, which this review aims to address. Therefore, this review contributes to the advancement of methods for spatially modeling diseases. The results of this systematic review offer a comprehensive understanding of modeling joint diseases, especially those transmitted by similar vectors, thereby aiding in their spread and mitigation. This study contributes to the methodology and practice of spatiotemporal modeling of emerging diseases. Its findings are of great significance to researchers and all sectors involved in epidemiological modeling and public health. Ultimately, this research seeks to enhance the understanding of joint modeling and spatiotemporal methods in health data applications to inform policies related to monitoring and surveillance. The objectives of this study was to systematically retrieve, summarize methods and examine the methods that have been used to jointly map diseases.
Methods: the results were reported according to the Preferred Reporting Item for Systematic Review and Meta-analysis guidelines [23]. Further the paper [11,15] on methodological review was used to guide the review writing.
Inclusion and exclusion criteria
Inclusion criteria: the study included Peer-reviewed studies utilizing joint spatial and spatiotemporal modeling techniques for disease mapping. Spatial models, defined by a geographical index, and temporal models, characterized by a time index. Spatiotemporal models, encompassing both geographical and time indices. Joint models addressing two or more diseases. Studies employing a minimum of two visualization or modeling techniques, with or without covariates, to assess diseases.
Exclusion criteria: studies employing only one visualization or modeling technique, with or without covariates, for disease assessment. Commentaries, expert reviews, or reports lacking original research, with only relevant research studies considered for inclusion.
Registration and protocol: this systematic review adheres to the guidelines outlined in the PRISMA statement. Additionally, the review has been registered on PROSPERO with the following reference number: CRD42021246889.
Search methods for identification of studies: a comprehensive search strategy was done for peer-reviewed articles that employ use of spatial, temporal, spatiotemporal and joint modelling techniques in diseases with no time and language limits. The following databases were searched: PubMed, Science direct, Scopus, Cochrane library, Trip database. Additional papers were identified by examining the reference lists of retrieved studies and by contacting the authors where necessary. A spatial model means that which has a geographical index, while a temporal model has a time index. Spatiotemporal implies those with both geographical index and time index, and the joint model means those that deal with two diseases. The literature search was confined to peer-reviewed journal articles published in English from January 2000 to June 2023. All search results were compiled, and duplicates were eliminated using EndNote. Initially, one author screened the titles and abstracts of the identified articles through keyword searches. Subsequently, papers meeting the inclusion criteria were subjected to a comprehensive evaluation by reading the full text. This latter stage was carried out independently by two authors. Any disagreements between the authors were resolved through discussion and consensus.
Study selection: two authors independently screened titles and abstracts to identify relevant studies for inclusion in the study. Studies that aligned with the research questions were further assessed for eligibility in a full-text review. Any discrepancies that arose were resolved through consensus or with the involvement of an independent arbitrator.
Data extraction and management: two reviewers independently used a standardized extraction form to gather data. This form was pre-tested and refined as needed. In cases of discordance, a third party, was consulted for input. The extraction tools contained essential information such as bibliographic details, research objectives, data sources, disease data, types of covariates, data analysis methods (including modeling approaches), and the generated results.
Assessment of risk of bias in included studies: two authors independently assessed all articles. Any disagreements or inconsistencies were resolved by an arbitrator. The critical appraisal was conducted using an adapted tool for assessing the quality and risk of bias in modeling studies, as described by Aswi et al., Wah et al. et Moher et al. [11,15,23]. Eight-point scoring criteria, modified to suit the aims, objectives, input data, model validity, results, and conclusions of individual studies, was employed. Screening questions and criteria guided the scoring process, with scores ranging from 0 (poor) to 2 (good) for each criterion. The overall quality of individual studies was categorized as very high (>13), high (11-13), medium (8-10), or low (<8) [23].
Handling missing data: any missing data were documented for consideration in the analysis. Efforts were made to collect missing information from the authors, and significant empirical data related to the population under analysis were analyzed.
Data synthesis: data from studies with similar covariates, diseases, and study designs were pooled. Due to variations in the measures used by the studies that met the inclusion criteria, the results were summarized in a narrative format. The interpretation of study findings took into account the methodological quality of the studies before concluding.
Sample size calculation: sample size calculation was not performed as all systematic reviews published during the search period and meeting the eligibility criteria were to be included.
Subgroup analysis: insufficient data prevented the conduct of subgroup analyses to explore the effects of covariates in modeling the diseases.
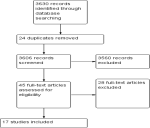
Literature search: a total of 3630 studies were retrieved from the various databases with 30 studies fully screened after the title and abstract review. Ultimately 17 studies were included for review and underwent quality assessment and synthesis (Figure 1).
Characteristics of included studies: the systematic review of infectious disease studies presents four distinct yet interconnected themes that encompass critical aspects of disease modeling, surveillance, and response. These themes collectively contribute to our understanding of infectious disease dynamics and the factors influencing their spread and impact. The four themes are listed below.
Surveillance and rapid response: this theme investigates into the strategies and approaches employed for the surveillance of infectious diseases, with a focus on swift and effective responses to emerging health threats. It explores how data collection, monitoring, and early detection play pivotal roles in disease control and public health interventions.
Disease emergence and environmental factors: understanding the emergence and re-emergence of infectious diseases is vital. This theme investigates the complex interplay between environmental factors, climate change, and disease dynamics. It highlights how shifts in environmental conditions can influence the prevalence, distribution, and emergence of infectious diseases.
Bayesian modeling and spatial analysis: Bayesian statistical modeling and spatial analysis techniques are at the core of this theme. It showcases the power of these methods in unraveling the complexities of disease modeling and spatial patterns. These approaches enable researchers to gain valuable insights into disease spread, clustering, and risk assessment.
Joint modeling of multiple diseases: investigating the joint distribution and interactions of multiple diseases is the focus of this theme. It demonstrates the significance of understanding how different diseases coexist and affect one another. Joint modeling provides a holistic view of disease dynamics, aiding in more effective disease management and control strategies. The themes from the table encompass a wide range of study sites, modeling approaches, and software tools used in infectious disease research. While some studies explicitly mention the use of covariates, others focus on Bayesian modeling, spatial analysis, and the impact of environmental factors on disease dynamics. These themes collectively contribute to our understanding of disease modeling, surveillance, and the factors influencing disease spread. The provided in the Annex 1 presents a summary of various studies related to infectious diseases, highlighting key details such as study objectives, study locations and periods, diseases involved, covariates, modeling approaches, key findings, further studies/recommendations, and software used.
Joint modeling of arbovirus
Most studies demonstrate various approaches to joint modeling, incorporating Bayesian methods, spatial analyses, and consideration of environmental and social factors to understand the complex dynamics of disease transmission, particularly in regions affected by multiple arboviruses [24-29]. Each study contributes insights that can inform public health strategies for surveillance, prevention, and control of these diseases. Martínez-Bello et al.[24] aimed to estimate the relative risk of dengue and Zika virus concurrently in Colombia from October 2015 to December 2016. Using Bayesian poisson joint models, the researchers found distinct risk distributions for Zika and dengue across different municipalities in Colombia, highlighting geographical variations in disease prevalence. Aguiar et al. [25] conducted a study in Brazil between 2015 and 2016, this study modeled potential outbreaks of Zika and chikungunya. Utilizing a statistical Maxent model, the research identified environmental and social conditions contributing to varying infection risks across Brazilian territories, emphasizing the impact of land use and other factors on disease ecology. Gómez-Rubio et al. [27] proposed a Bayesian hierarchical spatial-temporal model to jointly analyze multiple diseases in Spain. The research examined specific and shared spatial and temporal effects, the research aimed to pinpoint areas at high risk for various diseases, providing insights into disease dynamics useful for public health decision-making. Freitas et al. [28] focused on Rio de Janeiro, Brazil, during 2015 and 2016, this study utilized scan statistics analysis to explore the space-time dynamics of dengue, chikungunya, and Zika outbreaks. The findings highlighted clusters of these diseases and suggested targeted interventions in high-risk locations to enhance clinical management and vector-control measures. Schmidt et al. [29] conducted a study in Rio de Janeiro, Brazil, where dengue has been endemic, this study developed a poisson-multinomial spatial model to simultaneously analyze outbreaks of dengue, chikungunya, and Zika. By incorporating environmental factors, such as population density, the research assessed how these variables influence the spatial distribution and occurrence of different arboviruses. These studies underscore the importance of joint modeling approaches in understanding the complex epidemiology of arbovirus diseases, offering valuable insights into geographical variations, environmental influences, and potential strategies for disease surveillance and control.
Modeling approaches
The studies reviewed employ a range of sophisticated modeling approaches tailored to understand the epidemiology of arbovirus diseases across different regions and contexts. Commonly utilized modeling techniques include Bayesian approaches such as Bayesian Poisson joint models [24,28-31], Bayesian hierarchical spatiotemporal models [27,32], Bayesian change point models [16,17,33], multivariate negative binomial models [27,29], bivariate Poisson distributions [33,34], Geographic Information Systems (GIS)-based tools and geostatistical analyses [32] and scan statistics and space-time cluster analyses [28]. These methods allow researchers to integrate various data sources, account for spatial and temporal dependencies, and provide robust estimates of disease risk factors and transmission dynamics.
Additionally, multivariate negative binomial models and bivariate poisson distributions are used to analyze concurrent infections and mortality rates across multiple diseases, enhancing the accuracy of statistical inference compared to traditional univariate methods. Spatial cluster models implemented through reversible jump Markov chain Monte Carlo methods are also employed to detect joint and selective clustering patterns of diseases within specific geographic areas. Furthermore, Geographic Information System (GIS) tools and geostatistical analyses play a crucial role in spatial risk analysis, identifying environmental hotspots conducive to disease emergence and transmission. Scan statistics and space-time cluster analyses within R software are used to detect temporal and spatial patterns of disease outbreaks, guiding targeted interventions and public health measures. The diverse modeling techniques highlight the complexity of arboviral disease dynamics and underscore the importance of interdisciplinary approaches in epidemiological research to inform evidence-based public health policies and interventions.
Covariates used
Integrating covariates in disease studies enhances the understanding of disease dynamics and informs targeted interventions. The studies quantified the complex interactions between environmental, climatic, and sociodemographic factors, thereby improving public health strategies for disease prevention, surveillance, and control. Studies such as Aguiar et al. [25] looked at land use and urbanization Adegboye et al. [26] studied water bodies and irrigation. Ushijima et al. [32] looked on land use and geographic suitability while Freitas et al. [28] land use, urban planning and urbanization, socioeconomic status [29,31] looked urbanization, socioeconomic conditions for transmission of arbovirus. These factors influence vector habitats, breeding sites, and contact between humans and vectors. All these studies [25,26,28,31,35,36] considered climatic variables like temperature, rainfall, and seasonality. These factors affect vector biology, pathogen development, and the seasonal patterns of disease outbreaks. Held et al. et Souza-Santos et al. [37,38] focused on sociodemographic factors such as urbanization, socioeconomic status, and healthcare access. These factors influence exposure to disease, healthcare-seeking behavior, and the effectiveness of public health interventions.
The findings from studies incorporating environmental, climatic, and sociodemographic factors into disease modeling are crucial for advancing our understanding and prediction of disease dynamics. Environmental factors play a significant role in disease transmission dynamics [25,26,35,38]. These factors influence the distribution and abundance of disease vectors and reservoirs, impacting the spatial and temporal patterns of disease outbreaks. This is key in understanding the geographic distribution of disease risk and identify hotspots for targeted interventions.
Climatic variables such as temperature, rainfall, and seasonality significantly affect vector biology, pathogen development, and disease transmission. The studies [25-28,31,38] have demonstrated how these factors can influence disease incidence and outbreak dynamics. Incorporating climatic data into models assist in predicting seasonal peaks and troughs in disease incidence hence assessment of the potential impact of climate change on disease distribution and emergence. This can help in developing early warning systems based on climatic forecasts to enhance preparedness and response to disease outbreaks.
Sociodemographic factors, including population density, socioeconomic status, healthcare access, and cultural practices, shape vulnerability and resilience to disease. The studies [28,29,31] have underscored the importance of these factors in influencing disease transmission and response strategies. These behavioral aspects help to understand healthcare-seeking behavior, compliance with control measures, and community response to outbreaks. Thus, addressing disparities in disease burden and access to healthcare services across different demographic groups.
Incorporating environmental, climatic, and sociodemographic factors into disease modeling enhances our ability to predict, prevent, and control infectious diseases effectively. These findings are crucial for developing robust public health strategies that can mitigate disease risks and improve health outcomes globally.
Diseases involved and study sites
Both Zika and Dengue studies [3,24,25,31,32,38] are mosquito-borne viral infections transmitted primarily by Aedes mosquitoes. They pose significant public health threats in tropical and subtropical regions, causing outbreaks characterized by fever, rash, and in severe cases, neurological complications (Zika) and hemorrhagic fever (Dengue fever). The study was conducted in Colombia, Brazil, Thailand and Netherlands. Malaria and Cutaneous Leishmaniasis was studies in Afghanistan by Adegboye et al. [26]. Malaria is caused by Plasmodium parasites and transmitted through the bites of infected Anopheles mosquitoes. Cutaneous Leishmaniosis is caused by protozoan parasites of the Leishmania genus and transmitted through sandflies bites. Both diseases are endemic in Afghanistan, impacting rural populations with limited access to healthcare and vector control.
Multiple diseases were studied in Spain by Gómez-Rubio et al. [27]. Studying multiple diseases allows for a comprehensive understanding of disease interactions, co-occurrence patterns, and shared risk factors. This approach helps in developing integrated surveillance and control strategies to manage overlapping disease burdens effectively. Zoonotic Diseases in these studies [24,31,32], are infections transmitted between animals and humans. The research reads to understanding of the environmental and climatic factors influencing zoonotic disease transmission thus crucial for predicting and preventing outbreaks, especially in areas experiencing environmental changes and human-animal interaction.
Arbovirus diseases (Dengue, Chikungunya and Zika jointly) were studied in Brazil by the studies [25,28,29,38]. These arboviruses are viruses transmitted by arthropods (such as mosquitoes and ticks). Dengue, Chikungunya, and Zika viruses are major arboviral diseases affecting tropical and subtropical regions worldwide. These diseases have overlapping transmission cycles, and their co-occurrence presents challenges for disease control and management. Studying diseases in diverse geographic locations provides insights into regional differences in disease burden, transmission dynamics, and risk factors. This knowledge is crucial for tailoring interventions to local contexts and populations. Additionally, many of the diseases studied have global health implications due to their ability to spread rapidly across borders (e.g., Zika outbreaks in the Americas). Understanding their epidemiology and environmental drivers contributes to global health security and pandemic preparedness.
Studying diseases across various study sites and understanding their interactions with environmental, climatic, and sociodemographic factors is critical for advancing public health research, improving disease control strategies, and ultimately reducing the global burden of infectious diseases.
Quality assessment of included studies
The study adapted an assessment tool for modelling used by Aswi et al. [11] and attached in Annex 1 where the quality scores for reviewed paper ranged from 6 to 16 out of 16 (Annex 2). two studies were classified as low quality, three has high quality and 9 has high quality and two has very high quality The median score was 13/16 which is categorized as high quality as shown in result in Annex 3.
A variety of Bayesian spatial and spatiotemporal approaches were used in joint modelling of diseases [14,15,19,29,33,36,37]. Most studies adopted a fully Bayesian model with a spatially structured random effect using a Conditional Autoregressive (CAR) prior structure to investigate the relationship between the risk diseases and selected covariates spatial empirical Bayes smoothing was used for two studies to examine the spatial distribution of dengue and zika in Columbia [24,25]. Generalized Linear Mixed Models (GLMMs) with proper CAR spatial random effects were applied to develop disease maps, poisson temporal components were additionally incorporated, either as a temporal covariate [27,32]. Among the selected studies, only two studies used a GLMM with spatial, temporal and spatiotemporal random effects while one included these components along with an additional temporal covariate. Other GLMM spatiotemporal random effects models with incorporation of a temporal trend have also been developed. These studies [14,16,17,33] used a GLMM zero-inflated model. Alternative models included estimation of relative risk for the transmission of vector borne disease based on discrete time and space via a susceptible-infectious-recovered model for human populations.
Modeling covariates
The covariates used in models varied widely among the studies reviewed. Annex 3 shows two categories of covariates have been used widely i.e climatic covariates and migration. More than half of the studies used environmental/climatic factors in joint modelling of diseases. The climatic variables that were used are temperature and rainfall.
Implications for practice
Infectious diseases have spread in the globe faster and will be a big problem in the future. As most of these diseases are mosquito-borne diseases, it calls for careful study and understanding of the joint effect of these diseases for policy and containment. There is a need to continually review and validate models that help in mapping and surveillance of diseases jointly.
Implications for research
Modeling diseases jointly aids in prediction of the next disease outcome. Variables should be selected based on data variables and the effect they have on the whole model. The aim of variable selection should be to maximize predictions and accurate mapping of diseases. The spatiotemporal modeling approaches should incorporate the challenges facing the control and elimination of vector borne diseases like migration, and co-infection.
There are only a few proposed spatiotemporal models capable of jointly analyzing multiple diseases, highlighting the necessity for further model development in this area. This review comprehensively covers the approaches used in joint spatial and spatial-temporal modeling. The strategy employed in this review was transparent, as it encompassed all articles written in English. Relevant papers published in other languages were excluded, which aligns with the guidelines for reporting methodological reviews. The included studies offer a thorough description of vector-borne diseases and their potential for joint modeling. Consequently, conducting a meta-analysis in this study would not be appropriate.
What is known about this topic
- Bayesian spatial and spatiotemporal approaches are commonly used in joint disease modeling;
- Generalized Linear Mixed Models (GLMMs) are frequently employed for disease mapping, often incorporating spatial and temporal random effects;
- Covariates used in joint disease models vary widely, with environmental and climatic factors being the most common.
What this study adds
- This study systematically synthesizes methods for spatial and temporal modeling of diseases, identifying shared covariates across studies;
- It reviews factors influencing the development of joint disease models, providing insights into methodological advancements in disease surveillance;
- The review highlights the necessity for further model development in joint spatial and spatiotemporal modeling to address the limited number of proposed models capable of analyzing multiple diseases concurrently.
The authors declare no competing interests.
Samuel Mongare and Mr Lameck Agasa: conceptualization and designing of the systematic review. Draft the first manuscript: Leila Abdullahi: conducting the literature search, study selection and data extraction. Thomas Achia, Wycliffe Kipkirui and Antony Karanja: revising and drat critically for important intellectual content. All authors have read and agreed to the final manuscript.
The authors thank the African Institute for Development Policy (AFIDEP) for mentorship and training on systematic review.
Figure 1: characteristics of included studies
Annex 1: exploring joint modeling approaches for infectious diseases
Annex 2: risk of bias tool for assessment
Annex 3: risk of bias assessment for the study
- Wang RN, Zhang YC, Yu BT, He YT, Li B, Zhang YL. Spatio-temporal evolution and trend prediction of the incidence of Class B notifiable infectious diseases in China: a sample of statistical data from 2007 to 2020. BMC Public Health. 2022 Jun 17;22(1):1208. PubMed | Google Scholar
- Kuleshov Y, Wei Y, Inape K, Liu GJ. Spatio-temporal distribution of vector borne diseases in Australia and Papua New Guinea vis-à-vis climatic factors. J Vector Borne Dis. 2022 Apr-Jun;59(2):115-126. PubMed | Google Scholar
- Molina-Guzmán LP, Gutiérrez-Builes LA, Ríos-Osorio LA. Models of spatial analysis for vector-borne diseases studies: A systematic review. Vet World. 2022 Aug;15(8):1975-1989. PubMed | Google Scholar
- Real LA, Biek R. Spatial dynamics and genetics of infectious diseases on heterogeneous landscapes. J R Soc Interface. 2007 Oct 22;4(16):935-48. PubMed | Google Scholar
- Heesterbeek H, Anderson RM, Andreasen V, Bansal S, De Angelis D, Dye C et al. Modeling infectious disease dynamics in the complex landscape of global health. Science. 2015 Mar 13;347(6227):aaa4339. PubMed | Google Scholar
- Chowell G, Sattenspiel L, Bansal S, Viboud C. Mathematical models to characterize early epidemic growth: A review. Phys Life Rev. 2016 Sep;18:66-97. PubMed | Google Scholar
- Kreck M, Scholz E. Back to the Roots: A Discrete Kermack-McKendrick Model Adapted to Covid-19. Bull Math Biol. 2022 Feb 17;84(4):44. PubMed | Google Scholar
- Kretzschmar M. Disease modeling for public health: added value, challenges, and institutional constraints. J Public Health Policy. 2020 Mar;41(1):39-51. PubMed | Google Scholar
- Chen D, Moulin B, Wu J. Introduction to Analyzing and Modeling Spatial and Temporal Dynamics of Infectious Diseases. In book: Anlayzing and Modeling Spatial and Temporal Dynamics of Infectious Diseases (pp.3-18). Jones Wiley. December 2014. In press.
- Mahaki B, Mehrabi Y, Kavousi A, Schmid VJ. Joint Spatio-Temporal Shared Component Model with an Application in Iran Cancer Data. Asian Pac J Cancer Prev. 2018 Jun 25;19(6):1553-1560. PubMed | Google Scholar
- Aswi A, Cramb SM, Moraga P, Mengersen K. Bayesian spatial and spatio-temporal approaches to modelling dengue fever: a systematic review. Epidemiol Infect. 2018 Oct 29;147:e33. PubMed | Google Scholar
- Costa SDSB, Branco MDRFC, Aquino Junior J, Rodrigues ZMR, Queiroz RCS, Araujo AS et al. Spatial analysis of probable cases of dengue fever, chikungunya fever and zika virus infections in Maranhao State, Brazil. Rev Inst Med Trop Sao Paulo. 2018 Oct 25;60:e62. PubMed | Google Scholar
- Rodrigues M, Bonfim C, Portugal JL, Frias PG, Gurgel IG, Costa TR, Medeiros Z. Análise espacial da mortalidade infantil e adequação das informações vitais: uma proposta para definição de áreas prioritárias. Ciência & Saúde Coletiva. 2014;19:2047-54. PubMed | Google Scholar
- Knorr-Held L. Bayesian modelling of inseparable space-time variation in disease risk. Stat Med. 2000 Sep 15-30;19(17-18):2555-67. PubMed | Google Scholar
- Wah W, Ahern S, Earnest A. A systematic review of Bayesian spatial-temporal models on cancer incidence and mortality. Int J Public Health. 2020 Jun;65(5):673-682. PubMed | Google Scholar
- Knorr-Held L, Rasser G. Bayesian detection of clusters and discontinuities in disease maps. Biometrics. 2000 Mar;56(1):13-21. PubMed | Google Scholar
- Held L, Hofmann M, Höhle M, Schmid V. A two-component model for counts of infectious diseases. Biostatistics. 2006 Jul;7(3):422-37. PubMed
- Chidumwa G, Maposa I, Kowal P, Micklesfield LK, Ware LJ. Bivariate Joint Spatial Modeling to Identify Shared Risk Patterns of Hypertension and Diabetes in South Africa: Evidence from WHO SAGE South Africa Wave 2. Int J Environ Res Public Health. 2021 Jan 5;18(1):359. PubMed | Google Scholar
- Wakefield JC, Best NG, Waller L. 'Bayesian approaches to disease mapping', in Paul Elliott, and others (eds), Spatial Epidemiology: Methods and Applications (Oxford, 2001; online edn, Oxford Academic, 1 Sept. 2009). Accessed 23 July 2024.
- Tzougas G, di Cerchiara AP. Bivariate mixed Poisson regression models with varying dispersion. North American Actuarial Journal. 2023 Apr 3;27(2):211-41. Google Scholar
- Arnold BC, Ghosh I. On classical and Bayesian inference for bivariate Poisson conditionals distributions: theory, methods and applications. Communications in Statistics-Simulation and Computation. 2023 Nov 10:1-3. Google Scholar
- Amaral AVR, González JA, Moraga P. Spatio-temporal modeling of infectious diseases by integrating compartment and point process models. Stoch Environ Res Risk Assess. 2023;37(4):1519-1533. PubMed | Google Scholar
- Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015 Jan 1;4(1):1. PubMed | Google Scholar
- Martínez-Bello DA, López-Quílez A, Prieto AT. Joint Estimation of Relative Risk for Dengue and Zika Infections, Colombia, 2015-2016. Emerg Infect Dis. 2019 Jun;25(6):1118-1126. PubMed | Google Scholar
- Aguiar BS, Lorenz C, Virginio F, Suesdek L, Chiaravalloti-Neto F. Potential risks of Zika and chikungunya outbreaks in Brazil: A modeling study. Int J Infect Dis. 2018 May;70:20-29. PubMed | Google Scholar
- Adegboye OA, Al-Saghir M, Leung DH. Joint spatial time-series epidemiological analysis of malaria and cutaneous leishmaniasis infection. Epidemiol Infect. 2017 Mar;145(4):685-700. PubMed | Google Scholar
- Gómez-Rubio V, Palmí-Perales F, López-Abente G, Ramis-Prieto R, Fernández-Navarro P. Bayesian joint spatio-temporal analysis of multiple diseases. SORT-Statistics and Operations Research Transactions. 2019;43(1):51-74. Google Scholar
- Freitas LP, Cruz OG, Lowe R, Sá Carvalho M. Space-time dynamics of a triple epidemic: dengue, chikungunya and Zika clusters in the city of Rio de Janeiro. Proc Biol Sci. 2019 Oct 9;286(1912):20191867. PubMed | Google Scholar
- Schmidt AM, Freitas LP, Cruz OG, Carvalho MS. A Poisson-multinomial spatial model for simultaneous outbreaks with application to arboviral diseases. Stat Methods Med Res. 2022 Aug;31(8):1590-1602. PubMed | Google Scholar
- Rotejanaprasert C, Lawson AB, Iamsirithaworn S. Spatiotemporal multi-disease transmission dynamic measure for emerging diseases: an application to dengue and zika integrated surveillance in Thailand. BMC Med Res Methodol. 2019 Oct 26;19(1):200. PubMed | Google Scholar
- Gardini Sanches Palasio R, Marques Moralejo Bermudi P, Luiz de Lima Macedo F, Reis Santana LM, Chiaravalloti-Neto F. Zika, chikungunya and co-occurrence in Brazil: space-time clusters and associated environmental-socioeconomic factors. Sci Rep. 2023 Oct 21;13(1):18026. PubMed | Google Scholar
- Ushijima Y, Abe H, Nguema Ondo G, Bikangui R, Massinga Loembé M, Zadeh VR et al. Surveillance of the major pathogenic arboviruses of public health concern in Gabon, Central Africa: increased risk of West Nile virus and dengue virus infections. BMC Infect Dis. 2021 Mar 17;21(1):265. PubMed | Google Scholar
- Kim H, Sun D, Tsutakawa RK. A bivariate Bayes method for improving the estimates of mortality rates with a twofold conditional autoregressive model. Journal of the American Statistical association. 2001 Dec 1;96(456):1506-21. Google Scholar
- Esser HJ, Liefting Y, Ibáñez-Justicia A, van der Jeugd H, van Turnhout CAM et al. Spatial risk analysis for the introduction and circulation of six arboviruses in the Netherlands. Parasit Vectors. 2020 Sep 10;13(1):464. PubMed | Google Scholar
- Fone D, Hollinghurst S, Temple M, Round A, Lester N, Weightman A, Roberts K, Coyle E, Bevan G, Palmer S. Systematic review of the use and value of computer simulation modelling in population health and health care delivery. J Public Health Med. 2003 Dec;25(4):325-35. PubMed | Google Scholar
- Lawson AB, Zhou H. Spatial statistical modeling of disease outbreaks with particular reference to the UK foot and mouth disease (FMD) epidemic of 2001. Prev Vet Med. 2005 Oct 12;71(3-4):141-56. PubMed | Google Scholar
- Held L, Natário I, Fenton SE, Rue H, Becker N. Towards joint disease mapping. Stat Methods Med Res. 2005 Feb;14(1):61-82. PubMed | Google Scholar
- Souza-Santos R, Sobral A, Périssé ARS. High-risk spatial clusters for Zika, dengue, and chikungunya in Rio de Janeiro, Brazil. Rev Saude Publica. 2023 Jun 5;57:32. PubMed