Polymerase chain reaction fails to detect Listeria species from wild and cultured Nile tilapia (Oreochromis niloticus) caught from a large freshwater lake in Zambia
Prudence Mpundu, Marina Elisabeth Aspholm, Walter Muleya, Nawa Mukumbuta, John Bwalya Muma, Musso Munyeme
Corresponding author: Prudence Mpundu, Department of Environmental and Occupational Health, Levy Mwanawasa Medical University, Lusaka 33991, Zambia 
Received: 14 Sep 2022 - Accepted: 09 Nov 2022 - Published: 16 Nov 2022
Domain: Food safety
Keywords: Fish, Listeria species, Oreochromis niloticus, tilapia, Zambia
©Prudence Mpundu et al. PAMJ-One Health (ISSN: 2707-2800). This is an Open Access article distributed under the terms of the Creative Commons Attribution International 4.0 License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cite this article: Prudence Mpundu et al. Polymerase chain reaction fails to detect Listeria species from wild and cultured Nile tilapia (Oreochromis niloticus) caught from a large freshwater lake in Zambia. PAMJ-One Health. 2022;9:19. [doi: 10.11604/pamj-oh.2022.9.19.37355]
Available online at: https://www.one-health.panafrican-med-journal.com/content/article/9/19/full
Short communication 
Polymerase chain reaction fails to detect Listeria species from wild and cultured Nile tilapia (Oreochromis niloticus) caught from a large freshwater lake in Zambia
Polymerase chain reaction fails to detect Listeria species from wild and cultured Nile tilapia (Oreochromis niloticus) caught from a large freshwater lake in Zambia
![]() Prudence Mpundu1,2,&,
Prudence Mpundu1,2,&, ![]() Marina Elisabeth Aspholm3, Walter Muleya4, Nawa Mukumbuta5, John Bwalya Muma2,
Marina Elisabeth Aspholm3, Walter Muleya4, Nawa Mukumbuta5, John Bwalya Muma2, ![]() Musso Munyeme2
Musso Munyeme2
&Corresponding author
Fish have been identified as suitable vehicles for transmitting Listeria species to humans. Therefore, the thrust of this study was to determine possible Listeria species likely to circulate in freshly caught Nile tilapia (Oreochromis niloticus) in a large freshwater body. Accordingly, 150 Nile tilapia were sampled on landing. Of this number, half (n=75) were wild caught, while the other half (n=75) were from cage culture farms. All the sampled tilapia fish were independently caught from the same lake. Skin surfaces and gills were the only points sampled to restrict sites swabbed to those in contact with the environment. Swabs were processed using standard bacteriological culturing and identification tests. Only 2% (3/150) of the samples on culture and biochemical tests were presumptively Listeria. However, on Polymerase Chain Reaction (PCR), none yielded any noticeable results. None-detection of Listeria species on PCR may, in part, indicate the absence or undetectable levels in freshwater bodies. The detection of presumptive Listeria species phenotypically indicates possible contaminants other than Listeria. Our study has inherent limitations since it restricted itself to identifying contamination based on the fish's growth environment. We recommend determining the pathogen by incorporating enteric organs or whole fish analysis to increase isolation rates based on the current results.
Listeriosis is a severe foodborne disease caused by L. monocytogenes, a gram-positive bacterium belonging to the phylum firmicutes [1]. Fish and fish products contribute about 5.5% to 12.1% of confirmed listeriosis cases in the United States [2]. Previous studies [3-5] reported significant variations of 0.55% to 38% in the prevalence of Listeria species in raw fish samples collected from fish carcasses and fish markets in Nigeria, Iran, and India. In Africa, disease outbreaks linked to the consumption of specific foods are underreported, compared to water-related, because of inadequate water supply or treatment in many regions.
Most countries in the Southern part of Africa, including Zambia, do not differentiate between foodborne and waterborne diseases (outbreaks). L. monocytogenes is naturally present in the environment and tolerate a broad range of environmental stressors. In water and soil environments, L. monocytogenes integrates into multispecies biofilms. Its sturdiness allows L. monocytogenes to survive in refrigeration food production environments making it a significant food production concern. Raw fish contaminated with L. monocytogenes pose a public health concern mainly when inadequately heat treated or served as ready to eat food.
Listeria species identification involves culture methods based on selective enrichment followed by plating and characterization of single isolates. Rapid molecular tests, like Polymerase Chain Reaction (PCR), are available now. For instance, the housekeeping prs gene for the genus Listeria which encodes phosphoribosyl pyrophosphate synthase [6]. Although the prs gene does not differentiate different Listeria species, it nevertheless acts as an indicator of the genus Listeria in samples [6]. The most common tilapia species farmed in cages is Oreochrome niloticus. Due to its ease and cost-effectiveness, tilapia cage culture has boosted freshwater fish farming and economic interest in tropical countries. Tilapia is among the world's most frequently consumed fish types, and its farming improves food security, including income generation. Cultivation practices have raised concerns that pathogenic bacteria can contaminate the waters and confer a risk of foodborne disease. However, knowledge of the prevalence of Listeria species in tilapia caught from a large freshwater lake in Zambia is limited. Therefore, this study aimed to determine the presence of Listeria species circulating in freshly caught tilapia from a large freshwater lake in Zambia using PCR.
Study design: a cross-sectional study design was conducted between the periods of February to April 2021.
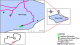
Study settings: Siavonga district is located along lake Kariba on the North shore in the Southern Province of Zambia, 16° 32' South of the equator and 28° 44' East. Most fish produced comes from aquaculture cage farms. The Siavonga district was chosen for the study due to its large number of aquaculture cage farms and capture fisheries. All the sites we selected for sampling the owners had to consent and fish availability during the study period. At capture, fisheries sampling was at three of the six recorded major sites along lake Kariba in the Siavonga district. These we coded as sites 1, 2, and 3. Out of 15 aquaculture cage farms in the Siavonga Town Council, only three gave consent for sampling. The coding for the sites was 4, 5, and 6 (Figure 1).
Sample size and sampling: sampling was done through the collection of fish to be sampled based on the two main fishing types found in the Siavonga district: aquaculture cage farms and capture fisheries. To get equal variation, we collected one hundred and fifty tilapia swabs from capture fisheries (n=75) and aquaculture cage farms (n=75). Swab samples from each fish came from the exterior surface and gills, independently sampled at the landing stage. We used random sampling at both the capture fisheries and the aquaculture cage farms to pick the individual tilapia for swabbing from the involved sites.
Sample collection and processing: we used a standard procedure for collecting swab samples from the fish, briefly, a sterile metal template to pre-mark a 5cm2 x 5cm2 area for swabbing. After swabbing the fish with a moist cotton swab, the swab was inserted back into a screw-cap tube containing Amies transport media. We recorded metadata such as farmed or wild-caught, collection site, and collection date. Sample storage was at -4°C before transportation, usually within 72hrs after sampling; we took it to the Microbiology laboratory at the School of the Veterinary Medicine University of Zambia. Samples were assigned laboratory codes upon receipt. Immediately after arrival, the swabs from the tubes were transferred into 9ml of pre-enriched broth (Oxoid) and incubated at 37°C for 48hrs. During the cultivation and identification of Listeria species, the L. monocytogenes strain ATCC19118 we used as a positive control.
Isolation and identification of Listeria species: the protocol for the isolation of Listeria species is previously described by Mpundu P et al. [7]. Briefly described as follows; firstly, a fish sample was inoculated into 9ml of pre-enriched broth (Oxoid) and incubated at 37°C for 24hrs. Later 1ml of pre-enriched broth (Oxoid) was transferred into 9ml of Fraser broth (Oxoid) (enriched with Listeria selective supplement) and vortexed for 1min, followed by incubation at 37°C for 48hrs. A loop full of the Listeria enriched culture was inoculated onto the surface of Listeria selective agar (Oxoid), incubated at 37°C for 48hrs, and observed for colonies with morphology typical for Listeria species, i.e., green-blue color, with a possible greenish sheen. Presumptive Listeria species colonies were sub-cultured on Nutrient Agar (Oxoid) incubated at 37°C for 24hrs to obtain pure colonies. The quality of the agar plates was tested by cultivating the L. monocytogenes strain ATCC19118. We performed standard biochemical tests on the purified presumptive Listeria species: Gram staining, catalase, sulphur indole motility, oxidase, urease, Voges Proskauer, and methyl-red tests.
Deoxyribonucleic acid (DNA) extraction and polymerase chain reaction identification of Listeria: polymerase chain reaction assays were performed on presumptive Listeria isolates to verify their identity. Chromosomal deoxyribonucleic (DNA) was isolated from the isolates grown on nutrient agar using the ZYMO Research quick DNA miniprep kit, following the manufacturer's instructions. For PCR detection of Listeria species, a partial 370bp fragment of the prs gene was targeted by using the primer pair prs -F (5'-GCT GAA GAG ATT GCG AAA GAA G-3') prs -R (5'-CAA AGA AAC CTT GGA TTT GCG G-3') [6]. PCR was under the following conditions initial denaturation at 94°C for 1 min, 30 cycles at 94°C for 45 s, 53°C for 45 s, 72°C for 2 min, and final extension step at 72°C for only 5 min. Finally, the PCR product was separated on a 1.5% agarose gel stained with ethidium bromide to visualize a potential 370 bp band. We used the L. monocytogenes strain ATCC19118 for quality control.
Data analysis: all the obtained data from the tilapia fish carcasses were entered in the excel datasheet and imported to Stata version 15 (Stata cop, College Station, Texas, U.S.A). The primary outcome was the presence of the Listeria species contamination from tilapia fish swabs. We calculated the prevalence of presumptive Listeria species contamination as the proportion of the total tilapia fish swabs collected that were contaminated phenotypically.
Ethics: approval of ethics was acquired from Excellence in Research Ethics Committee reference no. (Ref.no.2020-Jan-006). We sought permission from Siavonga Town Council and maintained the confidentiality of all sampling sites involved throughout this study.
Of the 150 tilapia samples collected from captured fisheries and aquaculture cage farms, 2.0% (3/150) were phenotypically indicative of Listeria species contamination. None of the presumptive Listeria isolates were prs positive. However, the positive control samples gave the expected 370 bp on the agarose gel, which secured reliability. The highest prevalence of presumptive Listeria isolates occurrence was found in samples from capture fisheries 2.7% (2/75) (95% CI: -1.048 - 6.453), aquaculture cage farms 1.3% (1/75) (95% CI: 1. 284 - 3.916) (Table 1).
The present study determined the prevalence of Listeria species contamination of tilapia from lake Kariba in the Siavonga district in Zambia. In the present study, none of the surface swab samples collected from fresh tilapia carcasses belonged to the genus Listeria on PCR. To the best of our proof, the present study is the first that investigated Listeria species contamination in tilapia from a large freshwater lake in Zambia. A similar study in India showed that Listeria species surface swabs had 72.4% of tropical fish and fishing environments [8]. The significant difference deduces from the differences in the sampled fish; Jeyasekaran and others collected sample swabs from fish markets and fish processing units. Onjong and others conducted a study on the microbiological safety of fresh tilapia in value chains, and a 2% prevalence of L. monocytogenes sampled at landing was recorded [9]. The differences in isolation with this present study deduction are from the sampled tilapia fish characteristics and techniques used in the isolation method. This present study utilized the prs gene to confirm the Listeria genus, a housekeeping gene found in all Listeria species. At the same time, Onjong and co-workers used VITEK 2 system version 0.8.01 biomerieux, Inc. Hazelwood, MO. These two methods may yield different results due to differences in sensitivity [10]. Further, this study only collected swabs at the landing stage without multiple processes.
Human activities occurring near sampled water bodies may thus influence the level of Listeria contamination in the fish. In the Siavonga district, cattle and crop farming are rare, and most people solely depend on fishing for a living. The low level of Listeria might partly be due to the lack of extensive farming activities in this area. The none-detection of Listeria contamination on PCR observed in the present study could also be explained because sampling was at the primary production stage. Furthermore, sampling was from a single lake, where environmental factors are assumed to be similar, previous studies reporting a higher prevalence of Listeria species mainly sampled fish from different water bodies.
Climatic conditions, mainly rainfall, contribute to bacterial contamination of surface water. The collection of samples was during the rainy season, when the probability of isolating Listeria species is usually higher than during the dry season, as reported in other studies. However, Hansen and others (2006) failed to find significant seasonal variations of L. monocytogenes levels in water from freshwater fish farms [11]. The principle limitation of this study is the low sample size. However, some studies have used less than ten (10) samples of fish from freshwater and have been able to isolate Listeria species. Additionally, the other limiting factor may link to the approach used. Including entire fish or the enteric contents would have increased the likelihood of isolating Listeria. On the other hand, large freshwater bodies have a practical dilution effect. Another insidious factor that we might have inadvertently ignored may have significantly affected the final results.
Results from the study intimate the likelihood of low levels of Listeria species in wild-captured and cultured tilapia from lake Kariba in the Siavonga district of Zambia. However, conducting further studies across different seasons is recommended. Additionally, there is need to repeat the same study with an increased sample size and inclusion of enteric organs and whole fish analysis to increase isolation rates.
What is known about this topic
- The prevalence of Listeria species in raw beef and poultry ranges from 0.095 to 15%.
What this study adds
- To the best of our awareness, this is the first study that investigated Listeria species contamination in tilapia from a large freshwater lake in Zambia.
The authors declare no competing interests.
Prudence Mpundu conceptualized the study and wrote the manuscript. Marina Elisabeth Aspholm and Musso Munyeme: validation of the molecular work protocol, and manuscript proofreading. John Bwalya Muma, Nawa Mukumbuta, and Walter Muleya contributed to manuscript editing and proofreading. All the authors have read and agreed to the final manuscript.
Table 1: presumptive Listeria species results in tilapia fish from captured fisheries and aquaculture cage farms from Kariba, a large freshwater lake, Zambia (n=150)
Figure 1: map of the Siavonga district showing the study areas
- Letchumanan V, Wong PC, Goh BH, Ming LC, Pusparajah P, Wong SH et al. A review on the characteristics, taxonomy and prevalence of Listeria monocytogenes. Prog Microb and Mol Bio. 2018 Dec 9;1(1). Google Scholar
- Dewey-Mattia D, Roberts V, Vieira A, Fullerton KE. Foodborne (1973-2013) and Waterborne (1971-2013) Disease Outbreaks - United States. MMWR Morb Mortal Wkly Rep. 2016 Oct 14;63(55):79-84. PubMed | Google Scholar
- Lennox JA, Etta PO, John GE, Henshaw EE. Prevalence of Listeria monocytogenes in fresh and raw fish, chicken and beef. J adv Microbial. 2017;3(4): 1-7. PubMed | Google Scholar
- Mashak Z. Banisharif F, Banisharif G, Reza Pourian M, Eskandari S, Seif A et al. Prevalence of Listeria species and serotyping of Listeria monocytogenes bacteria isolated from seafood samples. Egypt J Vet Sci. 2021;52(1):1-9. Google Scholar
- Menon KV, Sunil B, Latha C. Prevalence and antibiotic resistance profile of Listeria species associated with seafood's from fish catchment areas in Kerala, India. Vet World. 2021 Mar;14(3):777-783. PubMed | Google Scholar
- Doumith M, Buchrieser C, Glaser P, Jacquet C and Martin P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol. 2004 Aug;42(8):3819-22. PubMed | Google Scholar
- Mpundu P, Muma JB, Mukumbuta N, Mukubesa NM, Muleya W, Kapila P et al. Isolation, discrimination, and molecular detection of Listeria species from slaughtered cattle in Namwala District, Zambia. BMC Microbiol. 2022 Jun 18;22(1):160. PubMed | Google Scholar
- Jeyasekaran G, Karunasagar I, Karunasagar I. Incidence of Listeria spp. in tropical fish. Int J Food Microbiol. 1996 Aug;31(1-3):333-40 PubMed | Google Scholar
- Onjong HA, Ngayo MO, Mwaniki M, Wambui J, and Njage PMK. Microbiological safety of fresh tilapia (Oreochromis niloticus) from Kenyan fresh water fish value chains. J Food prot. 2018 Dec;81(12):1973-1981. PubMed | Google Scholar
- Martins KB, Ferreira AM, Mondelli AL, Rocchetti TT, LR de S da Cunha. Evaluation of MALDI-TOF VITEK® MS and VITEK® 2 system for the identification of Staphylococcus saprophyticus. Future Microbiol. 2018 Nov;13:1603-1609. PubMed | Google Scholar
- Hansen CH, Vogel BF and Gram L. Prevalence and survival of Listeria monocytogenes in Danish aquatic and fish-processing environments. J Food prot. 2006 Sep;69(9):2113-22. PubMed | Google Scholar