Antimicrobial resistant coliforms across four poultry production systems in Arusha and Moshi, Tanzania
Ruth Maganga, Emmanuel Sindiyo, Victor Moses Musyoki, Gabriel Shirima, Brian Maina, Blandina Theophil Mmbaga
Corresponding author: Ruth Maganga, The Nelson Mandela African Institution of Science and Technology, Arusha, Tanzania 
Received: 11 May 2021 - Accepted: 06 Jan 2022 - Published: 11 Jan 2022
Domain: Epidemiology,Microbiology,Molecular Biology
Keywords: Antimicrobial resistance, poultry, commensals, coliforms
©Ruth Maganga et al. PAMJ-One Health (ISSN: 2707-2800). This is an Open Access article distributed under the terms of the Creative Commons Attribution International 4.0 License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cite this article: Ruth Maganga et al. Antimicrobial resistant coliforms across four poultry production systems in Arusha and Moshi, Tanzania. PAMJ-One Health. 2022;7:4. [doi: 10.11604/pamj-oh.2022.7.4.29800]
Available online at: https://www.one-health.panafrican-med-journal.com/content/article/7/4/full
Research 
Antimicrobial resistant coliforms across four poultry production systems in Arusha and Moshi, Tanzania
Antimicrobial resistant coliforms across four poultry production systems in Arusha and Moshi, Tanzania
![]() Ruth Maganga1,2,3,&, Emmanuel Sindiyo1,
Ruth Maganga1,2,3,&, Emmanuel Sindiyo1, ![]() Victor Moses Musyoki4, Gabriel Shirima1, Brian Maina4,
Victor Moses Musyoki4, Gabriel Shirima1, Brian Maina4, ![]() Blandina Theophil Mmbaga3
Blandina Theophil Mmbaga3
&Corresponding author
Introduction: resistant to antimicrobials poses a threat to human and animal health. This study aimed to determine the prevalence of resistant coliforms in poultry cloacal samples collected from different poultry systems in Arusha and Moshi districts, Tanzania.
Methods: ten administrative wards were randomly chosen in Moshi and Arusha urban districts, with a random selection of one representative farm in each ward per production system (extensive, semi-intensive, intensive, and broiler systems). Per farm, 10 chickens were sampled using cloacal swabs. Samples were tested for the presence of coliforms using MacConkey agar without or with tetracycline, ciprofloxacin, ceftazidime, and Imipenem. R software was used for data analysis.
Results: of the 80 farms targeted, samples were collected from 79 farms representing a total of 746 samples, of which 648 (86.8%) had coliforms corresponding to 74 of the 79 sampled farms. There was no significant difference in the overall prevalence of coliforms between Moshi (86%) and Arusha districts (87%) (p=0.81). The overall proportions of resistant coliforms in Arusha and Moshi varied depending on each antimicrobial type. The prevalence of coliforms resistant to tetracycline (95%) across all farm types in both districts was higher compared to ciprofloxacin (72%), imipenem (71%), and ceftazidime (84%) (p<0.0001). The median counts of coliform resistance (in log cfu)) ranged from 4 to 10, with no significant distinctions between antimicrobial types.
Conclusion: there is a widespread presence of antimicrobial resistant coliforms in poultry production systems. High tetracycline resistance was observed across all farm types in both districts.
Antimicrobial resistance (AMR) is regarded as one of the major public health problems of the 21st century, causing more than 700,000 deaths worldwide each year [1]. According to Centers for Disease Control (CDC), more than 2.8 million antimicrobial-resistant infections occur in the United States per year, resulting in more than 35,000 deaths [2]. There is strong evidence of acquisition of AMR through food, between animals and humans, although the directionality has not been clearly established [3]. Food animals are important reservoirs of antimicrobial-resistant bacteria due to regular use of antimicrobials in animal production. There are numerous ecological niches in the food production chain, including many bacteria that coexist and undergo selection pressures continuously [4]. These bacteria can be transmitted directly or indirectly to humans through food consumption, contact with colonised, infected animals, animal products, or excreta such as urine, faeces, and blood [5]. Selective pressure due to consistent use of antimicrobials in animal production may lead to the acquisition of AMR genes in commensals through horizontal or vertical gene transfer [6]. Poultry, particularly chickens, are the world's most common and numerous species of livestock [7,8]. Indigenous chickens are widely distributed in rural and peri-urban areas, where they play an important role in income generation, food production, and social interactions [8-10]. In Tanzania, chickens are reared under various production systems, including scavenging (free-range and semi-intensive systems) and intensive systems that constitute indigenous breeds and broilers that are imported from other countries for consumption as meat [11]. The widespread use of antimicrobials in poultry farming to increase production and control diseases in chickens, particularly in semi-intensive, intensive and broiler systems, may select for antimicrobial resistant commensal organisms in chickens. Faecal coliforms are often considered as good indicator for selective pressure imposed by antimicrobial use and widespread in the chicken intestine [12]. Few reports exist on AMR in poultry production in East Africa [13]. To date, there has been no study in Tanzania that has investigated the prevalence of AMR in coliforms across all four poultry production systems. Existing research in Tanzania has either investigated prevalence of resistant bacteria in a single type of poultry farm [14], such as, extensive poultry systems, or compared between two poultry systems such as broiler and extensive systems [15]. Comparison of resistance patterns and prevalence across multiple regions and production systems is pertinent to gaining a deeper understanding of whether geographical differences and intensification of poultry production may impact AMR patterns and prevalence. The aim of this study was therefore to determine the prevalence of AMR in coliforms isolated from chickens in four distinct farm types; extensive, semi-intensive, intensive and broiler farm types.
Study design: a prospective cross-sectional study was designed to determine whether different poultry husbandry practices on different farm types are linked to varying degrees of antibiotic resistance in poultry populations at a given point in time between September 2016 and December 2017. The target number of samples was 800, consisting of 10 cloacal swabs per farm type (n = 4), per ward (n = 10) and per district (n = 2).
Study location and farm types: the study was conducted in urban Arusha and Moshi districts, northern Tanzania, and involved four production farm type systems: extensive, semi-intensive, intensive, and broiler. The farm types were categorized based on the degree of confinement of the chickens; supplementation of feed; use of veterinary services; labour; flock size and the number of poultry houses. Extensive systems had a flock size ranging from 5 to 50 indigenous birds kept under free range conditions and obtained food through scavenging around the homestead. These systems involved little input in terms of time management, provision of water, feeding, housing and disease control. The semi-intensive systems had 50 to 200 indigenous chickens enclosed in a facility but at some point, during the day the birds were released to scavenge. Commercial supplements were regularly incorporated into supplementary feed such as kitchen waste. Veterinary services were provided when necessary. Intensive systems were characterized as high-input urban and peri-urban commercial with 50 to 1000 birds reared for meat and egg production. The chickens were confined full time in constructed facilities and fed on feed and foundation stock from large-scale commercial poultry farms. The system involved full-time labour and the use of veterinary services for the prevention and management of the diseases. The broiler system focused on meat production by use of imported broiler breeds with flock sizes exceeding 200. The birds were confined full time in highly intensified units and involved the use of commercial feeds, supplements, and veterinary services.
Selection of wards, farms and chicken: we selected 18 wards from Arusha and 12 wards from Moshi containing all four production systems in a list of 25 and 21 administrative wards in Arusha and Moshi, respectively. Ten wards for each district were then randomly selected from these subsets. The selection was done by writing the name of each of the 18 wards in Arusha and 12 wards in Moshi on pieces of paper and folding to avoid disclosure and prevent bias during selection. Separately for Moshi and Arusha, the pieces of paper were randomized by tossing in a bowl. Then, five individuals each picked a piece of paper from the bowl without replacement. This procedure was done independently for Moshi and Arusha and repeated to generate a final list of 10 wards for each district. Four production systems were also randomly selected from a list of other similar farms within individual wards in a manner identical to the above-mentioned random selection process in each of the 10 wards. One farm was selected in each ward (randomly or purposively) per production system, followed by a convenience sampling of 10 birds in each farm. Random selection was carried out if a specific production system had more than 10 farms in a given ward. Each sampling day involved visiting one ward and sampling chickens through all four production systems. The selection of chicken in non-intensive production systems (i.e. extensive and semi-intensive) was carried out without taking into account the age of the chicken, whereas in intensive production systems (i.e. intensive and broiler systems) the selection was based on how the chicken were sorted in their cages by age. The majority of the farms separated chicken with an age gap of two weeks into different cages. In multi-cage farms we collected samples randomly from all the cages. For example, if a poultry farm belonging to a given system had three cages, three chickens would be picked out of each cage (making 9 samples) and the 10th chicken would be randomly selected from either cage.
Sample collection: of the 80 farms targeted for sample collection, only 79 farms were sampled. Consequently, 746 samples were collected. Cloacal swabs were collected using Amies transport media swabs (MML Diagnostics, Troutdale, OR) by gently swabbing the chicken cloaca mucosal wall. The swabs were transported to the Kilimanjaro Clinical Research Institute (KCRI) in ice-packed cool boxes and stored at -80°C in 1000ul mixture of 85% Brain Heart Infusion broth (Thermo Fisher Scientific Inc., Ottawa, ON, Canada) and 15% glycerol.
Isolation and enumeration of coliforms: the cloacal swabs were thawed at 2°C overnight, homogenized, and 50ul mixed with 450ul of maximum recover diluent (MRD) (Oxoid Thermo Fisher, Basingstoke, UK). The mixture was vortexed and 50ul plated using a spiral plater (Spiral System, Inc. Cincinnati, Ohio) on basic MacConkey agar and MacConkey agar supplemented with antimicrobials (tetracycline 16ug/ml, ciprofloxacin 4ug/ml, ceftazidime 8ug/ml, imipenem 4ug/ml), incubated for 24 hours at 37±3°C. Pink lactose fermenting colonies were isolated. Coliforms that grew on MacConkey agar with antimicrobials were considered resistant. Enumeration of coliforms was done using the spiral plater grid method on plain MacConkey agar and MacConkey with antimicrobials. A grid was placed on each plate, positioned on a level surface, and adjusted for the centre of each grid to match that of the plate on the viewer. The grid was divided into segments in which colonies were enumerated from the outer edge of the segment toward the centre allowing for an estimation of the corresponding microbial concentration (bacterial count/ml) on the entire plate according to standardised KCRI protocol.
Statistical analysis: data analysis was conducted using R (version 3.6.1). Chi-square test was used to determine any association between the presence of coliforms and district, farm type, and antimicrobial agents (tetracycline, ciprofloxacin, imipenem, and ceftazidime). The coefficient of determination (R2) was used to assess the strength of the relationship between counts of coliforms resistant to different pairs of antimicrobials. Kruskal-Wallis test was used to compare distributions of coliform counts between groups (farm type, district and antimicrobial type) and Mann-Whitney U test to compare medians.
Prevalence of coliforms: coliforms were present in 648 (86.8%) out of 746 samples collected (Table 1). There was no significant difference in the overall prevalence of coliforms between Moshi (86.4%) and Arusha districts (87.3%) (p=0.81). There was a difference between farm types in the prevalence of coliforms within the Arusha district (p<0.001) and within the Moshi district (p<0.01), but no consistent pattern in the prevalence across the farm types could be observed in either district. However, combining data across districts and across non-broiler farm types, showed that broiler farms had a significantly higher prevalence of coliforms (95.8%) than the other farm types combined (83.8%) (p<0.0001). Between districts, the extensive farm types showed the highest difference (of almost 11.5%), but this difference was not statistically significant (p=0.058). Resistance to each of the four antimicrobial types was detected in every extensive farm, broiler farm, and in 18 of 19 semi-intensive and intensive farms. There was no consistently higher prevalence of resistance in either district. Similarly, there was no consistent increase or decrease in the prevalence of resistant coliforms with the intensification of farm types (Table 2). The prevalence of tetracycline resistant coliforms across all farm types (95.0%) was higher compared to ciprofloxacin (71.5%), imipenem (70.81%), and ceftazidime (83.93%) (p<0.0001). The overall proportions of resistant coliforms in Arusha and Moshi varied depending on each antimicrobial type. In Arusha, there was a significant difference in the proportion of resistant coliforms between farm types for ciprofloxacin and imipenem (p<0.001) and ciprofloxacin, imipenem, and ceftazidime (p<0.01) in Moshi. There was evidence of interaction between farm type and district with the prevalence of ciprofloxacin-resistant coliforms; In Arusha, we noted a decline in the prevalence of resistant coliforms with the intensification of poultry production (with the exception of broiler farms) and an increase in prevalence with the intensification of poultry production in Moshi. The prevalence of imipenem-resistant coliforms in Moshi was higher, although only significant variation was noted in semi-intensive farms (p<0.001) while in both districts, no effect was observed on the prevalence of tetracycline, ciprofloxacin, and ceftazidime resistant coliforms (p>0.05) (p>0.05).
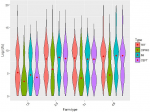
The median counts for tetracycline, imipenem, and ceftazidime resistant colonies were similar across the antimicrobial agents in the semi-intensive, intensive, and broiler farm types, while median resistance in the extensive system was lower in this group of antimicrobials. Median counts for ciprofloxacin resistant colonies were found to be higher in semi-intensive compared to extensive, intensive and broiler farmers (Figure 1). The median counts of coliform resistant to antimicrobials (in log (cfu)) ranged from 4 to 10, with no significant distinctions between antimicrobial agents. This pattern was also noted for individual antimicrobial types. Generally, there was a significant difference between districts for tetracycline, imipenem, and ceftazidime (p<0.001) and also in the distribution of coliform across all the four farm types (p<0.001). The distribution of total and resistant coliform counts was mostly bimodal and trimodal. In Arusha, for instance, bimodal distribution of coliform counts was observed in total coliforms and across all antimicrobial groups (i.e., tetracycline, ciprofloxacin, imipenem and ceftazidime resistant coliforms) in broiler farm types whereas in Moshi the distribution was predominantly trimodal across antimicrobial groups except for ciprofloxacin (Figure 2). Figure 3 illustrates the relationship between coliform counts (data transformed using log (cfu+1)) resistant to the four antimicrobial types. Variation was noted in the strength of relationship. Only 29% of the variation in coliform counts resistant to ciprofloxacin could be attributed to the change in the imipenem coliform counts (a weak association), whereas 55% of the variation in coliform counts resistant to ceftazidime could be explained by a change in the count of tetracycline resistant colonies (a strong association).
The present study demonstrates that chickens are reservoirs of antimicrobial resistant coliforms. Resistance to at least one of the four antimicrobials was observed in each farm. These findings reflect observations documented in other studies supporting the existence of antimicrobial resistant bacteria in poultry [14-16]. Furthermore, as an ultimate selective pressure for resistance, the use of antimicrobials in this region has been reported to be quite common among different animal keepers including poultry farmers of different ethnic groups [17,18]. Intensification of poultry farms did not significantly affect the prevalence of resistance. We noted there was no increase or decrease of resistant coliform with intensification of poultry farms. These findings undermine the assumption that intensification of poultry production increases the likelihood of higher prevalence. This is contrary to the inference made by a previous study in this setting, in which a higher prevalence was observed in intensive systems compared to extensive systems, although comparison was only conducted between two types of farms; commercial layer and free range [15]. The latter observation is corroborated by similar studies done in other parts of the world including Italy and Spain [19-23]. However, in concurrence to our findings, Obeng and colleagues in a similar study showed that there was no significant difference of resistant isolates between free range (extensive) and commercial chickens (intensive) [24].
Antimicrobials, in particular tetracycline, have been used in both districts for therapeutic and non-therapeutic purposes including prophylaxis and growth promotion [25]. Resistance to tetracycline was significantly higher in both districts for all farm types. This was anticipated as resistance to tetracycline in animals was found to be quite common in previous studies in the northern zone of Tanzania [11-18] and other parts of the world such as Egypt, the United States, Portugal, and Norway [16-29]. Extensive usage of tetracycline could be linked to its accessibility, low price, wide spectrum, and long shelf life. These factors, combined with the propensity of tetracycline resistance genes to co-select with other types of resistance genes, function to promote widespread tetracycline resistance [30,31].
Although the prevalence of resistance to other antimicrobials such as imipenem, ciprofloxacin, and 3rd generation cephalosporin was present, it was significantly lower than resistance to tetracycline. Resistance varied between farm types, and this may have been associated with the differential use of antimicrobials within those farm types. Resistance to imipenem and ceftazidime was not expected, as these antibiotics are not commonly used in poultry production [11]. Currently, there is no information on imipenem usage in poultry production documented in these districts to help illustrate the cause of this resistance. Nonetheless, this remains an area of concern to explore other potential driving factors besides antimicrobial use. Conversely, cephalosporin resistance reservoirs have been identified in water sources including tap and open water sources that are widely used in poultry production within these districts [32].
Our study found a strong association in a number of coliforms that were resistant to specific antimicrobial types; tetracycline versus that of ceftazidime; tetracycline versus imipenem; imipenem versus ceftazidime. Although such findings do not support the existence of numerous forms of resistance in a single isolate, a strong association suggests the likelihood of co-selective pressure between different antimicrobials. This potential is well known for tetracycline, often aided by co-transfer of tetracycline resistance genes along with genes responsible for conferring resistance to other forms of antimicrobials in the same genetic elements [30-33]. Tetracycline has been found present in large plasmids with several resistance genes in other studies, proposing that tetracycline has a high potential for co-selection with other genes.
Bimodal or trimodal distribution of bacterial populations have been linked to the presence of diverse resistance mechanisms [34,35]. In the present study, the distribution of coliform counts differed significantly between samples, and bimodal and trimodal distributions were observed [36,37]. Microbes can develop novel mechanisms when exposed to sub-therapeutic levels of antimicrobials, leading to lethal selection. Under sub-therapeutic conditions, microorganisms may accumulate multiple resistance mutations in a step-by-step process, contributing to heterogeneity in bacterial populations and the evolution of resistance with minimal effects [38]. Alternatively, in some bacterial populations, bimodal distributions may be prompted by phenotypic switching (often slow growth). In the event of ecological or antimicrobial resistance stress, it is presumed that there is a subset of bacterial species that defaults to slow growth, dividing bacterial populations into fast and slow growers. These slow-growing cells are referred to as persister cells and are thought to be capable of maintaining this phenotype for a long period of time [39-41]. Genetically, similar or different bacterial species may therefore respond heterogeneously to antimicrobial treatment, producing multimodal distributions [39-41]. The major limitation in this study was that some farms had fewer than 10 chickens, and therefore, future research may consider using a large sample size.
There is a widespread presence of resistant coliforms in poultry production systems in the two districts in Northern Tanzania. In both Moshi and Arusha, high resistance to tetracycline was observed across the farm types. Additionally, no consistent antimicrobial resistance pattern changed with the gradation of the farm types. Considering imipenem resistance in poultry production systems, prospective studies need to examine all factors influencing resistance in the one health continuum. In order to reduce the development of antimicrobial resistant bacteria in animal production, poultry keepers should receive adequate antimicrobial resistance stewardship training in order to promote good practices and reduce antimicrobial overuse in poultry farms. Furthermore, continuous surveillance of antimicrobial resistance patterns in poultry should be conducted to detect emergent resistance patterns, especially those of last resort significance, such as third-generation cephalosporins and imipenem, which could have major implications for human medicine.
Funding: the project was funded by the Biotechnology and Biological Sciences Research Council (BBSRC). This work was supported by the Zoonoses and Emerging Livestock Systems (ZELS), University of Glasgow College of Medical, Veterinary and Life Sciences, Kilimanjaro Clinical Research Institute and The Nelson Mandela African Institution of Science and Technology (NM-AIST).
What is known about this topic
- Food animals are important reservoirs of antimicrobial-resistant bacteria due to regular use of antimicrobials in animal production;
- Indiscriminate use of antimicrobials in animal husbandry leads to increased prevalence of antimicrobial resistance.
What this study adds
- A high prevalence of tetracyline resistance was found in all four poultry productions;
- Resistance to antimicrobial agents was found in all four poultry production systems;
- There was no consistent increase or decrease in prevalence of antimicrobial resistant coliforms with intensification of farm types.
The authors declare no competing interests.
RM, ES, GS and BTM were all involved in the project's conceptualization and design. RM and ES did sample collection, performed the laboratory experiments and analyzed the data. RM, VM and BM prepared the manuscript. All the authors read and approved the final manuscript.
The authors would like to acknowledge all members of the University of Glasgow Research into Bacteria Infectious Diseases (ORHBID) laboratory. The authors would also like to acknowledge the Kilimanjaro Research Institute's zoonosis laboratory and the director therewith. Our appreciation also goes to the research assistants and laboratory technicians who participated in sample analysis.
Table 1: cloacal swab samples positive for coliforms (%) across four farm types in Arusha and Moshi districts
Table 2: distribution of antimicrobial resistant coliforms by antimicrobial and farm type in Arusha and Moshi districts
Figure 1: distribution of antimicrobial resistant coliforms by antimicrobial type within farm types across the two districts
Figure 2: density plots for the total and resistant coliform counts (in log cfu)) in cloacal swabs across the four antimicrobials types
Figure 3: pair wise relationships between coliform counts (data transformed using log (cfu+1) resistant to the four antimicrobial types
- O´Neill Jim. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. London. Review on Antimicrobial Resistance. 2014.
- Center for Disease Control and Prevention. Antibiotic resistance threats in the United States. Atlanta, GA: US Department of Health and Human Services. 2019.
- Muloi Dishon, Ward MJ, Pedersen AB, Fevre EM, Woolhouse ME, van Bunnik BA. Are food animals responsible for transfer of antimicrobial-resistant Escherichia coli or their resistance determinants to human populations? a systematic review. Foodborne Pathog Dis. 2018;15(8):467-474. PubMed | Google Scholar
- Acar JF, Moulin Guy. Antimicrobial resistance at farm level. Rev Sci Instrum. 2006;25(2):775-792. PubMed | Google Scholar
- Chuang KH, Whitney-Miller CL, Chu CY, Zhou Zhongren, Dokus MK Schmit, Shannon Barry CT. MicroRNA-494 is a master epigenetic regulator of multiple invasion-suppressor micrornas by targeting ten eleven translocation 1 in invasive human hepatocellular carcinoma tumors. Hepatology. 2015;62(2):466-480. PubMed | Google Scholar
- Hammerum AM, Heuer OE. Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin Infect Dis. 2009; 48(7):916-921. PubMed | Google Scholar
- Perry BD. Investing in animal health research to alleviate poverty. Int Liv Res Inst. 2002;1:1-140. Google Scholar
- Moreki JC, Dikeme Richard, Poroga B. The role of village poultry in food security and HIV/AIDS mitigation in Chobe District of Botswana. Liv Res for Rural Dev. 2010;22(3):1-7. Google Scholar
- Minga UM, Mtambo MMA, Katule AM, Mutayoba SK, Mwalusanya NA, Lawrence PP et al. Improving the health and productivity of the rural chicken in Africa: research and development efforts in Tanzania. In Aciar proceedings. 2001;1998:134-139. Google Scholar
- Thornton Philip. Mapping poverty and livestock in the developing world. Int Liv Res Inst. 2002;1:1-118. Google Scholar
- Sindiyo Emmanuel, Maganga Ruth, Thomas KM, Benschop Jackie, Swai Emmanuel, Shirima Gabriel et al. Food safety, health management, and biosecurity characteristics of poultry farms in arusha city, northern Tanzania, along a gradient of intensification. East African Heal Res Journal. 2018;2(2):168-80. PubMed | Google Scholar
- Alekshun MN, Levy SB. Commensals upon us. Biochem. Pharmacol. 2006;71(7):893-900. PubMed | Google Scholar
- Hakanen JJ, Seppälä Piia, Peeters MC. High job demands, still engaged and not burned out? The role of job crafting. Int J Behav Med. 2017;24(4):619-27. PubMed | Google Scholar
- Hamisi Zainabu, Tuntufye Huruma, Shahada Francis. Antimicrobial resistance phenotypes of Escherichia coli isolated from tropical free range chickens. Int J Sci Res. 2014;3(9):34-37. PubMed | Google Scholar
- Rugumisa Bernadether, Call DR, Mwanyika GO, Subbiah Murugan, Buza Joram. Comparison of the prevalence of antibiotic-resistant Escherichia coli isolates from commercial-layer and free-range chickens in Arusha district, Tanzania. Afr J Microbiol Res. 2016;10(34):1422-1429. Google Scholar
- Katakweba AA, Muhairwa AP, Lupindu AM, Damborg Peter, Rosenkrantz JT, Minga UM et al. First report on a randomized investigation of antimicrobial resistance in fecal indicator bacteria from Livestock, Poultry, and humans in Tanzania. Microb Drug Resist. 2018;24(3):260-268. PubMed | Google Scholar
- Caudell MA, Quinlan MB, Subbiah M, Call DR, Roulette CJ, Roulette JW et al. Antimicrobial use and veterinary care among agro-pastoralists in Northern Tanzania. PLoS One. 2017;12(1):e0170328. PubMed | Google Scholar
- Caudell MA, Mair Colette, Subbiah Murugan, Matthews Louise, Quinlan RJ, Quinlan MB et al. Identification of risk factors associated with carriage of resistant Escherichia coli in three culturally diverse ethnic groups in Tanzania: a biological and socioeconomic analysis. Lancet Planet Heal. 2018;2(11):e489-e497. PubMed | Google Scholar
- Pesciaroli Michele, Magistrali CF, Filippini Giovanni, Epifanio EM, Lovito Carmela, Marchi Lucia et al. Antibiotic-resistant commensal Escherichia coli are less frequently isolated from poultry raised using non-conventional management systems than from conventional broiler. Int J Food Microbiol. 2020;314:108391. PubMed | Google Scholar
- Much Peter, Sun Hao, Lassnig Heim, Koeberl-Jelovcan Sandra, Schliessnig Herald, Stueger HP. Differences in antimicrobial resistance of commensal Escherichia coli isolated from caecal contents of organically and conventionally raised broilers in Austria, 2010-2014 and 2016. Prev Vet Med. 2019;171(1):104755. PubMed | Google Scholar
- Álvarez-Fernández Elena, Cancelo Amaya, Díaz-Vega Carmen, Capita Rosa, Alonso-Calleja Carlos. Antimicrobial resistance in E. coli isolates from conventionally and organically reared poultry: a comparison of agar disc diffusion and Sensi Test Gram-negative methods. Food Control. 2013;30(1):227-234. Google Scholar
- Musa Laura, Pratrizia CP, Branciari Raffaella, Menchetti Laura, Bellucci Sara, Ranucci David et al. Antimicrobial susceptibility of Escherichia coli and ESBL-producing Escherichia coli diffusion in conventional, organic and antibiotic-free meat chickens at slaughter Animals. 2020;10(7):1215. Google Scholar
- Miranda JM, Vazquez BI, Fente CA, Calo-Mata Pilar, Cepeda Alberto, Franco CM. Comparison of antimicrobial resistance in Escherichia coli, Staphylococcus aureus, and Listeria monocytogenes strains isolated from organic and conventional poultry meat. J food Prot. 2008;71(12):2537-42. PubMed | Google Scholar
- Obeng AS, Rickard Heather, Ndi Olasumbo, Sexton Margaret, Barton Mary. Antibiotic resistance, phylogenetic grouping and virulence potential of Escherichia coli isolated from the faeces of intensively farmed and free range poultry. Vet Microbiol. 2012;154(3-4):305-315. PubMed | Google Scholar
- Dahshan Hesham, Abd-Elall AMM, Megahed AM, Abd-El-Kader MA, Nabawy EE. Veterinary antibiotic resistance, residues, and ecological risks in environmental samples obtained from poultry farms, Egypt. Environ Monit Assess. 2015;187(2):1-2. PubMed | Google Scholar
- Fahrenfeld Nicole, Knowlton Katharine, Krometis LA, Hession WC, Xia Kang, Lipscomb Emily et al. Effect of manure application on abundance of antibiotic resistance genes and their attenuation rates in soil: field-scale mass balance approach. Environ Sci Technol. 2014;48(5):2643-2650. PubMed | Google Scholar
- Tacão Marta, Moura Alexandra, Correia Antonio, Henriques Isabel. Co-resistance to different classes of antibiotics among ESBL-producers from aquatic systems. Water Res. 2014;48:100-107. Google Scholar
- Chee-Sanford JC, Aminov RI, Krapac IJ, Garrigues-Jeanjean Natalie, Mackie RI. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl Environ Microbiol. 2001;67(4):1494-502. PubMed | Google Scholar
- Sunde Marianne, Norström Madelaine. The prevalence of, associations between and conjugal transfer of antibiotic resistance genes in Escherichia coli isolated from Norwegian meat and meat products. J Antimicrob Chemother. 2006;58(4):741-747. PubMed | Google Scholar
- Al-Ghamdi Mastour, Al-Mustafa ZH, El-Morsy FE, Al-Faky A, Haider I, Essa H. Residues of tetracycline compounds in poultry products in the eastern province of Saudi Arabia. Public Health. 2000;114(4):300-304. PubMed | Google Scholar
- Harada Kazuki, Asai Tetsuo. Role of antimicrobial selective pressure and secondary factors on antimicrobial resistance prevalence in Escherichia coli from food-producing animals in Japan. J Biomed Biotechnol. 2010;2010:180682. PubMed | Google Scholar
- Lyimo BM, Buza Joram, Woutrina Smith, Subbiah Murugan, Call DR. Surface waters in northern Tanzania harbor fecal coliform and antibiotic resistant Salmonella spp capable of horizontal gene transfer. African J Microbiol Res. 2016;10(11):348-356. Google Scholar
- Choi JM, Woo GJ. Transfer of tetracycline resistance genes with aggregation substance in food-borne Enterococcus faecalis. Curr Microbiol. 2015;70(4):476-484. PubMed | Google Scholar
- Shah Devang, Zhang Zhigang, Khodursky AB, Kaldalu Niilo, Kurg Kurg, Lewis Kim. Persisters: a distinct physiological state of E coli. BMC Microbiol. 2006 Jun 12;6:53. PubMed | Google Scholar
- Smith Peter, Christofilogiannis, Panos. Application of normalised resistance interpretation to the detection of multiple low-level resistance in strains of Vibrio anguillarum obtained from Greek fish farms. Aquaculture. 2007;272(1-4):223-230. Google Scholar
- Mazzariol Annarita, Tokue Yutaka, Kanegawa TM, Cornaglia Giuseppe, Nikaido Hiroshi. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob Agents Chemother. 2000;44(12):3441-3. PubMed | Google Scholar
- Mcmurry LM, Oethinger Margret, Levy SB. Over expression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol Lett. 1998;166(2):305-309. PubMed | Google Scholar
- Wistrand-Yuen Erik, Knopp Michael, Hjort Karin, Koskiniemi Sanna, Berg OG, Andersson DI. Evolution of high-level resistance during low-level antibiotic exposure. Nat Commun. 2018;9(1):1599. PubMed | Google Scholar
- Kussell Edo, Kishony Roy, Balaban NQ, Leibler Stanislas. Bacterial persistence: a model of survival in changing environments. Genetics. 2005;169(4):1807-14. PubMed | Google Scholar
- Balaban NQ, Merrin Jack, Chait Remy, Kowalik Lukasz, Leibler Stanislas. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622-1625. PubMed | Google Scholar
- Radzikowski JL, Vedelaar Silke, Siegel David, Ortega ÁD, Schmidt Alexander, Heinemann Matthias. Bacterial persistence is an active σS stress response to metabolic flux limitation. Mol Syst Biol. 2016;12(9):882. PubMed | Google Scholar