Site specific bacterial load, enterobacterial occurrence and antibiotic susceptibility patterns in Nile Tilapia (Oreochromis niloticus), water and sediment from earthen aquaculture ponds
Gordon Takop Nchanji, Andrielle Larissa Tchamba Kemajou, Bertrand Tatsinkou Fossi, Nancielle Mbiatong Tchatat, Manuel Ritter, Samuel Wanji
Corresponding author: Bertrand Tatsinkou Fossi, Department of Microbiology and Parasitology, University of Buea, P.O. Box 63, Buea, Cameroon 
Received: 05 May 2021 - Accepted: 12 Jan 2022 - Published: 04 Feb 2022
Domain: Microbiology, Food safety
Keywords: Aquaculture, tilapia, antibiotic resistance, enterobacteria, THAPC, Escherichia coli, Enterobacter cloacae
©Gordon Takop Nchanji et al. PAMJ-One Health (ISSN: 2707-2800). This is an Open Access article distributed under the terms of the Creative Commons Attribution International 4.0 License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cite this article: Gordon Takop Nchanji et al. Site specific bacterial load, enterobacterial occurrence and antibiotic susceptibility patterns in Nile Tilapia (Oreochromis niloticus), water and sediment from earthen aquaculture ponds. PAMJ-One Health. 2022;7:16. [doi: 10.11604/pamj-oh.2022.7.16.29693]
Available online at: https://www.one-health.panafrican-med-journal.com/content/article/7/16/full
Research 
Site specific bacterial load, enterobacterial occurrence and antibiotic susceptibility patterns in Nile Tilapia (Oreochromis niloticus), water and sediment from earthen aquaculture ponds
Site specific bacterial load, enterobacterial occurrence and antibiotic susceptibility patterns in Nile Tilapia (Oreochromis niloticus), water and sediment from earthen aquaculture ponds
![]() Gordon Takop Nchanji1,2, Andrielle Larissa Tchamba Kemajou1,2,
Gordon Takop Nchanji1,2, Andrielle Larissa Tchamba Kemajou1,2, ![]() Bertrand Tatsinkou Fossi1,2,&, Nancielle Mbiatong Tchatat1,2,
Bertrand Tatsinkou Fossi1,2,&, Nancielle Mbiatong Tchatat1,2, ![]() Manuel Ritter3, Samuel Wanji1,2
Manuel Ritter3, Samuel Wanji1,2
&Corresponding author
Introduction: aquaculture is the most rapidly growing food production sector. The intensive nature of freshwater fish aquaculture is associated with practices which predispose aquaculture produce to bacterial contamination. To prevent losses due to bacterial diseases, there has been an increase in the uncontrolled use of antibiotics, a good prerequisite to the development of antibiotic resistance - a major global problem. Hence, we aimed at investigating the enterobacterial diversity and occurrence of antimicrobial resistance in Nile Tilapia and its earth pond environment.
Methods: fish samples, pond water and sediments were collected from four earthen ponds located at Yato, Cameroon, in three different collection trips for Total Heterotrophic Aerobic Plate Count (THAPC) and enterobacterial isolation. Bacterial characterisation and antibiotic sensitivity were assessed using the API 20E kit and the standard Kirby-Bauer disc diffusion method, respectively.
Results: THAPC ranged from 1.0x102 to 3.0x109 CFU/mL across samples and within each pond, bacterial loads were highest in fish parts and lowest in the surrounding environment. Sixteen enteric genera and 24 species were identified, with Escherichia coli (16.49%) and Enterobacter cloacae (12.37%) being most abundant, especially in the gills and intestines. The highest degree of resistance was observed with Amoxicillin and Ampicillin, with more than 60% of tested isolates being resistant. Multiple drug resistance was also observed in all sample types, ponds and collection time points, and highest in fish parts.
Conclusion: this study re-emphasizes the potential of freshwater fish aquaculture as carrier of human pathogenic and multiple drug resistant bacteria.
Aquaculture, which presently accounts for about 50% of the global fish production for food is the most rapidly growing animal production sector [1], and because of increased marine pollution, overfishing and global climate change, the push has been towards a rapid transition from species capture to aquaculture production model [2]. Fish is one of the preferred sources of cheap and healthy proteins in developing countries [3], such as Cameroon - where fish farming has been reported to be a profitable business [4]. Freshwater aquaculture is still an emerging practice in Cameroon with great potential since the gap between demand and production is huge. The effect of this huge demand for fish has led to shift towards intensive freshwater fish aquaculture, characterized by several risks factors that lead to microbial contamination of fish [5]. These include manipulation of breeding cycles, high stocking density of fishes in pond and the feeding or fertilisation of ponds with agricultural by-products including animal droppings, imposing a certain amount of stress on the fish and increasing the predisposition of fish to bacterial contamination.
Several studies have reported on pathogenic bacteria associated with fish in aquaculture systems [6-9], and some of which are human pathogens [10,11]. In an attempt to promote growth, prevent or treat bacterial infections in intensive aquaculture setups, antimicrobials have been massively administered to fish. This increases the risk of emergence and spread of resistant bacteria capable of causing infection in both animals and humans as there is no discrimination between the classes of therapeutic antimicrobials used in human and food producing animal [12]. Moreover, fish do not effectively metabolize these antibiotics hence 70 - 80% of drugs fed to the fish are excreted in the aquaculture environment [13]. These unmetabolized drugs subsequently contaminate products meant for human consumption as well as the human handlers [14]. These drugs are also environmentally very persistent, thus exert selective pressure on the microbiota of aquaculture ecosystem [15]. Due to the rise in antibiotic resistance, there is increasing recognition that urgent action is needed to avoid inappropriate use of antibiotics in animal husbandry and aquaculture, as well as humans to maintain low resistance levels and prevent the proliferation of bacteria resistant to certain drugs [16].
Additionally, antimicrobial resistance patterns observed in inland animal husbandry have also been observed in aquaculture [17]. Tilapia is one of the most grown freshwater fish species in the world and the most cultivated species in Cameroon. This species is environmentally resilient, capable of resisting harsh temperatures, low dissolved oxygen and heavy stocking density, which exposes tilapia to bacterial contamination and encourage antimicrobial abuse. In light of the fact that there is a drive by the government to increase aquaculture production nationwide towards addressing national demand, this study was aimed at investigating the enterobacterial diversity of Nile Tilapia, from earthen ponds and to assess the occurrence of antibiotic resistant bacteria of food safety importance in earth ponds that might greatly impact public health and economic sectors.
Pond conditions: four earthen ponds located at Yato, Littoral Region of Cameroon, along the banks of the River Mungo (9° 33' 30" N, 4° 03' 02" E) were chosen for the study. Around the Mungo bridge, the river is about 150-200 m wide and has a depth of 5-15 m [18] and the soil type consists mainly of sandstone, intercalated with limestone and shale [19]. The site belongs to the Equatorial Monsoon climatic zone, characterized by annual rainfall ranging from 2000 to 10000 mm with an mean annual temperature of 21 ± 2.2°C [20]. The main activities in this region are artisanal in-channel sand mining, fishing, water supply, oil palm and banana farming [18]. These ponds are opened-air rectangular earth ponds 8-10 metres apart, surrounded by vegetation, palm trees, banana plants and fruit trees. The ponds are fed with waste from poultry, piggery, and palm oil production. These ponds established at the banks of River Mungo are not operated under flow-through systems and must be refilled about 3 times every 2 weeks with water from River Mungo to compensate for evaporation and seepage.
Sample collection: samples of tilapia, pond water and sediments were purposively collected from four earthen ponds. Nile Tilapia was fished out of the pond using a fishing line and a bait (earthworms or chicken intestine). Harvested fish samples were placed into transparent containers with pond water. A total of 52 tilapia were sampled from 4 ponds (Pond 1 to 4) within three collection trips; April (C1), December 2016 (C2) and May 2017 (C3). Twelve fish samples were collected at first trip and 20 each during the second and third trips, respectively. Fish samples from each pond were collected into separate containers. Six water samples were collected at different positions around each pond ~20 cm below the water surface into 60 mL sterile plastic collection cups. Using a handmade sediment sampler [21], six sediment samples were also collected at the same areas of the pond where water samples were collected. The containers were appropriately labelled and then placed in between ice packs for transportation to the laboratory within an hour of collection.
Sample processing and analysis: upon arrival at the laboratory, live fish were humanely killed through a sharp blow to the head. The skin of each fish was passively washed by pouring sterile water over the skin surface of the fish to wash off any transient bacteria before processing. A labelled sterile cotton swab was used to swab both sides of the fish, avoiding the opercular and anal regions, to collect the mucosal bacteria population on the surface of the fish's skin. Each fish sample was then aseptically weighed, its length measured and recorded. The fish skin was then disinfected with 70% alcohol and dissected to expose the gills and intestine of the fish. Aseptically, one gram each of gill and intestine was weighed and ground separately from each fish sample. The skin swab, ground gill and intestine were homogenised into 9 mL of 0.1% sterile peptone water and serially diluted (10-fold) to the 10-8 dilution. The six water and sediment samples collected per pond were divided into two, and three water/sediment samples were mixed to generate a composite sample, giving two composite water and sediment samples per pond. Therefore, each pond had 2 composite sediment samples and 2 composite water samples. One gram of each composite sediment sample was weighed and homogenised into 9 mL of sterile 0.1% peptone water. One mL of each composite water sample was also homogenised with 9 mL of sterile 0.1% peptone water. The homogenised water and sediment samples were serially diluted (10-fold).
Bacterial culture and microbial loads: total Heterotrophic Aerobic Plate Count (THAPC) was used to assess the total microbial loads in each sample by spreading 0.1 mL from each dilution on nutrient agar. To isolate enteric bacteria, each sample was also streaked on Eosine Methylene Blue (EMB) agar (Liofilchem, srl, Roseto, Italy) and Salmonella Shigella (SS) agar (Liofilchem). These plates were incubated for 24 hours at 30°C. For THAPC plates, the number of colonies were counted and recorded. The isolated bacteria from EMB and SS agar plates were sub-cultured to obtain pure colonies that were then stored in nutrient agar stabs at 4°C for 2-4 weeks prior to characterisation. All media were prepared according to manufacturers' instructions and cultured using standard methods.
Phenotypic characterization of isolates: gram staining was done on enteric bacterial pure isolates from nutrient agar and oxidase tests were performed using oxidase test strips (Liofilchem). Catalase tests were performed using 3% hydrogen peroxide (Gilbert Laboratories, Caen, France). Cells from pure cultures were grown on nutrient agar (Liofilchem) at 30°C, re-suspended in 0.85% NaCl API medium (BioMérieux, Inc., Marcy l'Etoile, France), and later inoculated into API 20 E strips (BioMérieux, Inc.) for additional phenotypic testing and characterisation. The test procedure was performed following the manufacturer´s instruction. The strips were incubated at 35°C, and results were recorded at 24h. Characterised isolates were then stored in 20% glycerol.
Antibiotic susceptibility testing: the phenotypically characterised isolates were then subjected to antibiotic susceptibility testing using the standard Kirby-Bauer disc diffusion method [22] as recommended by the Clinical and Laboratory Standards Institute [23]. Susceptibility to 11 antibiotics which belong to the major antibiotic classes (the penicillins, cephalosporincs, macrolide, aminoglycosides, carbapenams, fluroquinolone, sulphonamides, tetracyclines and nitrofurantoin) were tested. The antibiotics investigated were sulfamethoxazole and trimethoprim (23.75 μg/1.25 μg), gentamicin (10 μg), cefuroxime (30 μg), ceftriaxone (30 μg), imipenem (10 μg), tetracycline (30 μg), ampicillin (10 μg) and ofloxacin (5 μg) all obtained from BD BBLTM, USA. Erythromycin (15 μg), nitrofurantoin (100 μg) and amoxicillin (10 μg) were obtained from Abtek, UK. Each plate was incubated at 30°C for 24 hours, followed by the measurement of the zones of inhibition using a transparent ruler.
Statistical analysis: data collected was entered into Microsoft Excel 2013 datasheets and analysed using both Microsoft Excel and GraphPad Prism 8.0.2 (GraphPad Software, San Diego, USA). Microsoft Excel was used to generate Log10 of bacterial loads initially calculated and expressed as CFU/mL or CFU/g. Hypothesis investigated was the difference between mean microbial loads across collection trips, ponds and sample units (skin, gills, intestines water and sediment). To test for normality of the data, the Shapiro-Wilk test was employed. For normally distributed data, the one-way ANOVA was used to test this hypothesis and the Turkeys multiple comparison test was employed to test which means differed significantly from the others when the one-way ANOVA was significant. Wherever the number of groups were less than 3, the unpaired t-test was used to test the difference among means. For data that did not pass the Shapiro-Wilk test, the Kruskal-Wallis test was used to test the difference between groups and when significant, the Dunn's multiple comparisons test was used to determine which means differed significantly from the others. In the event where the Dunn's multiple comparisons test showed no difference despite a significant Kruskal-Wallis test, a Mann-Whitney test was used to compare pairs of datasets. Where the number of groups for comparison equaled 2, the Mann-Whitney test was used to test for differences. Statistical significance was set at P<0.05.
Experimental fish: a total of 52 fish samples were collected from the four ponds with a total of 13 tilapia per pond. Their weights ranged from 29.3g to 278.7g, with the lengths ranging from 11.2cm to 25.9cm (Table 1). Since none of the sampled fish presented with any skin abnormality and no fish mortality was observed in any of the ponds during sampling, the fish samples were considered as healthy.
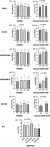
Bacterial loads: in general, the THAPC recorded ranged from 1.0 x 102 to 3.0 x109 CFU/mL. When the bacterial loads of each sample type were considered across ponds (Figure 1 A,B,C,D,E), THAPC did not differ significantly between the sample units across the ponds except for the gills (p=0.04) (Figure 1 B). When considered across sample collection rounds (Figure 1 F,G,H,I,J), bacterial loads differed significantly between collection time points for skin (p=0.028) (Figure 1 F), gills (p=0.0012) (Figure 1 G), intestines (p=0.0004) (Figure 1 H), pond sediment (p<0.0001;) (Figure 1 I), and pond water (p=0.0001) (Figure 1 J). Considering the pooled THAPC picture irrespective of collection trip or pond, the bacterial loads differed significantly among sample units and was significantly highest in fish intestines and lowest in water and sediment (p<0.0001;) (Figure 1 K). When pooled per pond (Figure 1 A,B,C,D) or per collection trip (Figure 1 E,F,G), the bacterial loads differed significantly among sample units in all ponds, highest in fish parts and lowest in the surrounding environment.
Biochemical characterization: biochemical and API characterisation of the presumptive bacteria isolates revealed the presence of 22 enteric genera and 43 species, with Escherichia coli being the most abundant (16.49%), followed by Enterobacter cloacae (12.37%). The bacteria species identified included: Acinetobacter spp, Aeromonas hydrophila, Aeromonas salmonicida, Escherichia coli, Enterobacter cloacae, Plesiomonas shigelloides, Citrobacter freundii, Edwardsiella tarda, Erwinia spp, Klebsellia pneumonia, Pseudomonas fluorescens, Raoultella ornithinolytica and Serratia liquefaciens (Table 1). The API database did not identify 3/194 of the isolates. All the ponds were contaminated and among the samples, the intestines, followed by the skin and gills were the most contaminated. The most abundant characterised isolate from the intestinal samples was Escherichia coli (20%) and Enterobacter cloacae was most abundant in the gills (23.4%) and skin (15.1%) respectively (Table 1).
Antimicrobial susceptibility: the antibiotics used in this study belong to the following classes of antibiotics; aminoglycosides, macrolides, penicillins/cephalosporins, quinolones, sulfonamide and tetracycline, which are the most commonly used classes of antibiotics in animal production and human medicine. The highest degree of resistance was observed with the penicillins amoxicillin and ampicillin, with 72% and 65.5% of tested isolates respectively, being resistant to these two antibiotics. Resistance was also high for Erythromycin as 58.5% of isolates tested were resistant. Gentamicin and ofloxacin inhibited most of the isolates (Figure 2). This general picture of resistance profile was also presented as per ponds and therein a similar trend was observed across the four ponds for each antibiotic (Figure 2 Aa). When presented across collection time points, resistance increased in the third phase for the drugs with highest resistance (AMX, AM and E) but reduced for the drugs with least resistance (IPM, CN and OFX) (Figure 2 Bb). Multiple drug resistant (MDR) species were also identified and presented as per their site of occurrence (Figure 3 A), per pond (Figure 3 B) and per collection time point ( Figure 3 C). Two of the most prevalent resistant profiles (E and AMX-AM-IPM) were present in all five sample types, 2 (AMX-AM-E and AMX-AM-E-SXT-TE) in four samples, while 2 were found only in fish samples (AMX-AM-CRO and AMX-AM-E-F) (Figure 3 A), highlighting that all combinations of obtained MDR isolates can be found in fish and thus, can affect humans through the consumption of fish. The three most prevalent resistant profiles (AMX-AM-E, E and AMX-AM-IPM) were present in all ponds while 8 of the most prevalent profiles (AMX-AM-CRO, AMX-AM-E-F, AMX-AM, AMX-AM, E-SXT-TE and AMX-AM-CXM-F) where found in 3 of four ponds (Figure 3 B). For the distribution of MDR across collection time points, the 3 most prevalent MDR profiles were found in all 3 collections while the next two most prevalent were recorded only in the 3rd collection trip. Seven of most frequent MDR profiles were found in 2 of 3 collection trips (Figure 3 C).
The quantitative and qualitative assessment of the bacterial loads, enterobacterial microbiota and their antibiotic resistance profile of the Nile Tilapia and its freshwater earth pond environment from Yato is revealed to be quite diversified. From our observation, this is the first study which seeks to investigate the presence and antimicrobial resistance profile of enterobacterial species found in tilapia fish and its freshwater aquaculture earth pond environment in Cameroon. The fish gills, skin and intestines, and the pond water and sediment constituted the samples, and the results demonstrate the diversity of enterobacteria across the different sample categories investigated. The variation in loads and species observed in this study is quite similar to results obtained by Pakingking [9] and Al-Harbi and Uddin [24], when investigating bacterial microbiota in pond water and sediment, fish gills and intestines of tilapia that was cultured in earthen ponds in the tropical country of the Philippines and brackish waters in Saudi Arabia, respectively. Generally, loads were higher in the intestines compared to the other tested samples, and lowest in the pond environment. The observed difference could be because most of the organisms from the environment accumulate in the intestines followed by gills. It was also observed that the loads are higher in the warmer season and this was probably because of higher temperatures which increase microbial metabolism.
Looking at the pooled microbial loads by pond, it was generally discovered that the loads between sample types (fish skin, gills and intestines, and pond water and sediment) differed significantly (P<0.5) (Figure 1 A,B,C,D). When each sample type was pooled per collection (Figure 1 E,F,G), the loads between samples were still statistical different (P<0.5). Bacterial loads in the fish were generally significantly higher than the loads in the environment and this is in accordance with other authors. In all, when all the loads were pooled irrespective of collection trip or pond, the loads were highest in intestines, followed by skin, gills and lowest in the environment (P<0.0001) (Figure 1 K). This is similar to trends reported by Kaktcham et al. [10] and Al-Harbi and Uddin [24], who performed similar studies in earth ponds and brackish water but slightly different from that reported by Pakingking et al. [9] in that the loads were higher in gills.
The bacterial count of the skin samples ranged from 105 - 108 CFU/g and is slightly lower compared to mean count of 109 CFU/g reported by Shinkafi and Ukwaja [25] in skin of tilapia sold in the market. The higher count in the study by Shinkafi and Ukwaja [25] could be as a result of post-harvest handling and storage since these tilapia samples were collected from the market. The load in the skin is slightly higher than that in the surrounding water and this is similar to what is reported by [26] in common carp ponds in India where similarity was reported between the loads in the pond water and skin of the common carp fish samples.
A closer look at each sample type across collection trips and across ponds revealed a better picture of the variation in bacterial loads. Looking at the loads when pooled across ponds (Figure 1 A), there was no statistical difference among skin loads across the four ponds. This clearly shows that there is no variation in bacterial loads across ponds at each collection time point, indicating that population of bacteria on the skin are more the same constant at same temperature. The pooled bacterial load of the skin differed between the three collection phases (P=0.028) (Figure 1 F). These loads where significantly highest in the warmer months of December and is most likely due to the increase microbial activity due to higher temperatures.
Taking a look at the tilapia gill samples, the THAPC of the gill samples ranged from 104 - 108 log CFU/g and was higher than that reported by Parkingking et al. [9] and Al-Harbi and Uddin [24] who reported counts of 105 - 107 CFU/g and 106 CFU/g respectively. The counts across ponds and collection phases of the intestines ranged from 105 - 109 CFU/g. This was slightly less than the total heterotrophic bacteria count range of 103 - 108 CFU/g, 107 - 108 CFU/g and 109 - 1010 CFU/g reported by Pakingking et al. [9] and by Kaktcham et al. [10] respectively. The higher loads might be as a result of the accumulation of microbes from the environment as water filters through the gills and during feeding of the fish with waste from agriculture and animal husbandry. The pooled THAPC count of the gills was statistically different across ponds, being highest in pond 2 (P=0.04) (Figure 1 B). When pooled per collection round, there was also a significant difference among counts on gills across the three collection trips (P=0.0012) (Figure 1 G), highest during the warmer month of collection 2. The loads being different across ponds and collection trip, imply that factors other than temperature contribute to the variation and the loads in the gills vary much more than that observed in the skin which is influenced largely by temperature of surrounding pond water.
The bacterial load of the intestines when pooled did not differ across ponds (Figure 1 C) but were significantly different across collection round and significantly highest in both second and third collection phases (P<0.0004;) ( Figure 1 H). This difference in pattern where the highest loads were not only higher in collection trip 2 is indicative of the fact that temperature might be less influential on the intestinal loads as compared to that of the skin. It is important to note that the similarity in loads in intestines across ponds but the difference across collection time points might be indicative of the implication of temperature or practices in increasing or decreasing loads.
The THAPC of the sediment samples ranged from 102 - 108 CFU/g and as expected, were higher than that obtained for the pond water. The THAPC count in sediments reported in this study is similar to the range of 105 - 106 and 106 - 107 CFU/g reported by Thanh et al. [27] and Al-Harbi and Uddin [24] in sediments of earth and brackish water ponds, but higher than those reported by Pakingking et al. [9] who reported counts ranging from 103 - 105 CFU/g in sediments of earth ponds. Kaktcham et al. [10] working in freshwater earthen ponds in the Western Highlands of Cameroon reported even higher counts of 109 - 1010 CFU/g in sediments, higher than what we obtained in the coastal region around the Mungo River. In addition to pond fertilization and manipulation practices, the soil type can account for the differences between both studies.
When pooled, there was no difference among bacterial loads of sediments across the four ponds (Figure 1 D) and this indicates that the sediment population is similar across ponds at each collection time point. Considering the collection phases, the pooled bacterial load of the pond sediment was different between the three collection phases (p<0.0001;) (Figure 1 I). Strikingly, the loads were highest in the third collection trip, implying that the difference in loads were not mainly influenced by temperature, but surely other factors which favour the accumulation of bacteria in the sediments.
Irrespective of collection phase or pond, the THAPC count of the water samples ranged from 103 - 107 CFU/mL. Our findings are similar to the THAPC range of 103 - 104 CFU/mL obtained by both Pakingking et al. [9] and Al-Harbi and Uddin [24] who investigated loads in water of earthen ponds in the Philippines and brackish water in Saudi Arabia respectively, but less than that of 109 - 1010 CFU/ml reported in Cameroon by Kaktcham et al. [10] in the Western Highlands of Cameroon. Since these are all tropical regions with generally similar climatic conditions, the differences between our result and that of Kaktcham et al. [10] can therefore be as a result of differences in the quality of source water, pond fertilization and manipulation practices across study sites. The bacterial load of the surrounding pond water was similar across ponds and did not differ between ponds when pooled (Figure 1 E). When considered as per collection round, the loads were significantly different across collection trips and lowest in the first strip (P=0.0001) (Figure 1 J). It is important to note that the water samples during second collection were lost as the containers leaked out.
Generally, it was expected that bacterial loads will be higher during the second collection which was in the warmer month of December compared to those of the first or third sample collection. This was observed for skin and gills samples and is in line with results obtained by Karimi [28] who worked with tilapia from freshwater aquaculture farms and the Mosinga Dam in Kenya. The author attributed the higher bacteria loads in the dry season compared to the wet season to increased bacteria growth at higher temperatures. Markosova and Jezek [29], in a 6-year long study, equally reported a similar trend with an increase in the load of indicator bacteria in carp fish ponds with increasing water temperature as they reported highest loads in the summer months.
It is also important to note that the variation in bacteria load across ponds for skin and pond water was negligible compared to that among gills, intestines and sediment samples. This may be because of the inability of these two sample categories to accumulate organisms, thus maintaining stable bacterial loads across the ponds. When looking at the variation at each collection period and across collection periods, it was realized that only the loads of the gills differed significantly at each collection and across collection trips. The reason for this is not very clear but may probable be evidence of more influence of environmental parameters on the gills more than any other sample type. Considering the bacterial loads across sample types per pond, it was observed that the loads were different across sample types, being significantly higher in the fish and lower in the environmental pond water and sediment. Some researchers have postulated that the microbiota of the fish is a reflection of the environment and according to Pakingking et al. [9], the bacterial quantity and quality of the rearing environment of tilapia is a mirror image of the bacterial quantity and quality of the tilapia gills and intestines. The findings do not give enough evidence to corroborate such suggestion since this study reported significantly lower loads and genera in the pond environment than in the fish. There were 10 genera present only in fish tissues, while 6 genera were found only in pond water or sediment. This could suggest that the fish microenvironment supports the propagation of some bacteria that cannot flourish in the environmental pond water or sediment. Given that no study is without potential limitations, it is important to highlight that a further description of the biological, physical or chemical characteristics of the sample units (fish, water and sediment), could have given other opportunities for analysis and discussion of the measured outcomes (loads and isolates).
Phenotypic characterization of the isolates revealed that bacteria isolated during this study are similar to enteric bacteria reported by other authors in fish or aquaculture environment. The bacteria identified using the API 20E kit belonged to 22 gram negative bacteria genera and 41 species. The genera identified included Acinetobacter, Aeromonas, Citrobacter, Enterobacter, Erwinia, Escherichia, Morganella, Plesiomonas, Pseudomonas, Edwardsiella, Raoultella, Salmonella, Serratia, Pasteurella and Vibrio (Table 1). These findings are in partial accordance to the study on the bacteria flora of Nile Tilapia cultured in brackish water by Pakingking et al. [9]. Even though [9] documented a similar number of bacteria genera, the absence of Escherichia coli from their work is a major difference with our study which strikingly was the most abundant bacteria isolated from the Nile Tilapia in earth pond aquaculture farms at Yato, Mungo. This high prevalence of E. coli in our study can have its source from handlers during pond fertilization practices using waste from chicken and pig farming, and palm oil extraction or from the source water which is collected from River Mungo. Given that the river is highly used for economic activities of in-channel sand mining and fishing which exposes the river to contamination by the human pathogen E. coli.
Karimi [28], working with tilapia harvested from freshwater fish ponds and the Masinga Dam also reported on bacteria genera similar to those we reported in our study, notably; Acinetobacter, Aeromonas, Citrobacter, Escherichia, Edwardsiella, Enterobacter, Klebsiella, Plesiomonas, Pseudomonas, Salmonella and Vibrio. There are also some similarities with the study on Nile Tilapia from four lakes in Northern Cameroon by Donkeng et al. [30] and another study carried out by Musefiu et al. [31] on Catfish and Nile Tilapia in Nigeria. Akoachere et al. [32] in their investigation of bacterial species associated with various species of fish from the coastal regions of Cameroon, also reported the presence of Escherichia, Citrobacter, Enterobacter Serratia, and Klebsiella which was observed in this study. It is important to point out that in their study, Escherichia coli was also the most abundant bacteria they reported.
Amongst the bacteria identified were species pathogenic to human especially members of the coliform group with Escherichia coli being the most abundant. In addition to being the most abundant species, only Escherichia was present in all sample types. It is important to mention that this is an important human pathogenic species which is of significant public health concern. Other important pathogenic species that have been reported include Pseudomonas aeruginosa and Klebsiellia pneumonia. Kaktcham et al. [10] also reported on presence of human pathogenic genera of public health interest in earth ponds including Escherichia, Salmonella and Vibrio which was also observed in this study. The findings from our study, similar to that from [10] and Akoachere et al. [32] clearly underline the fact that these fish samples are a clear potential health hazard to humans, be it handlers or consumers. In this light, there has been evidence of transmission of fishborne resistant bacteria to humans [33] even though most often, genetic evidence fails to link many of these suspected piscine zoonoses to human infections [34]. Besides being a potential source of human infection, these species can also contribute in the transfer of antibiotic resistance from aquaculture bacteria to human pathogens, even those from non-aquatic environments which widen the public health hazard that can result directly or indirectly from the aquaculture environment. Many studies have raised this call for concern concerning antibiotic resistance in the environment and food systems that can affect humans [35,36] and it is thus clear that it is of prime importance to stop the use of antibiotics in food animals [16]. The antibiotic susceptibility assessment of the bacteria microbiota of the Nile Tilapia from Yato, along the River Mungo revealed that they were mostly resistant to the penicillin class of drugs i.e. Amoxicillin and Ampicillin, followed by the macrolide Erythromycin, while highest susceptibility was to the fluroquinolone, ofloxacin and the aminoglycoside Gentamycin (Figure 2). The high resistance to penicillins is in line with the report by Donkeng et al. [30], and Dib et al. [37] who reported the increasing environmental incidence of resistance to beta-lactam antibiotics. It is worth pointing out that these macrolide and penicillin drugs form part of the critically important antimicrobials for human medicine due to their high frequency of use in human medicine and also form part of the drug classes with limited available therapies to treat serious bacterial infections in people [38].
It is important to underline that our study focuses on earthen pond aquaculture which is practiced by approximately 83.3% of the fish farmers in the country, with 39.2% of which use animal manure (chicken droppings (20.5%) and pig dung (18.7%) to fertilize the ponds [39]. Looking at the most prevalent resistant profiles overall, resistance to the penicillin-macrolides profile was highest and this is similar to reports by Akinbowale et al. [40] and Kathleen et al. [41], who arrived at similar conclusions while working with bacteria isolated from aquaculture sources in Australia and Borneo. This resistance profile was followed by macrolides only, penicillins-carbapenams and penicillins-cephalosporins-nitrofurantoins in frequency of occurrence. In all, this trend was spread across all four ponds (Figure 2 A) and collection time points (Figure 2 B). It was observed that the more prevalent multiple drug resistance profiles were spread across fish and pond environment samples (Figure 3 A), across ponds (Figure 3 B) and across collection time points (Figure 3 C), except for the AMX-AM-CRO and AMX-AM-E-F resistance profiles which were reported only in fish samples but not in pond environment (Figure 3 A). This indicates a circulation in the pond system of MDR species underlining the importance of this for veterinary and public health because of the risk this poses to both fish and humans.
Taking a closer look at the most prevalent genera, the E. coli strains were not of a prevalent resistance phenotype, as opposed to the Aeromonas, Enterobacter, Klebsiella and Pleisomonas genera. The AMX-AM-E resistance profile occurred most in the Enterobacter and Klebsiella genera, the AMX-AM-IPM phenotype was more present in the genus Aeromonas, and the AMX-AM occurred most in the genera Pleisomonas. Aeromonas hydrophilia was resistant to both ampicillin and tetracycline and this is in line with findings by [42]. Lee and Wendy [42] also reported on the high resistance of Edwardsiella tarda to ampicillin and tetracycline which differs from our findings as we report no resistance in our study. It is worth highlighting that in contrast to reports by Martins et al. [6], who reported Pleisomonas as a high carrier of tetracycline resistance, we did not observe resistance to tetracycline by Pleisomonas species. This may suggest that the Plesiomonas species in our study sites may have acquired resistance genes from other organisms. This demonstrates further the health risk that is posed by these organisms found in earth pond aquaculture systems.
Our results clearly show the resident bacteria and their antimicrobial resistance patterns in the skin, intestines and gills of tilapia, and the rearing environment (pond water and sediment). Moreover, bacteria residing in the tilapia and its rearing environment consist of genera of opportunistic and/or fish pathogens and human pathogenic species. Therefore, the need for good aquaculture practices of tilapia and its pond environment cannot be overemphasized. Our findings and reports from other authors [14,43,44] further emphasize the importance of this call for concern about antimicrobial use and antimicrobial resistance organisms in the environment including the aquaculture environment. Follow-up studies will be needed to clarify the mechanisms of resistance, how they are transferred, their relationship with fish feed and animal droppings added to the ponds, and investigate alternatives to antibiotics that can be suitable in the tilapia earth pond setting.
What is known about this topic
- Specific enteric bacteria have been reported in aquacultured fish or environment;
- The excessive use of antibiotics in food production systems has worsened the threat of antimicrobial resistance in both animal and human health.
What this study adds
- The occurrence of enterobacterial species in earth pond aquaculture setting of importance to human and animal health;
- The resistance to antibiotics of isolated strains is high with occurrence of multiple drug resistance profiles detected in all ponds, collection time points and pond environment.
The authors declare no competing interests.
Gordon Takop Nchanji: field activities (equal); methodology (equal); formal analysis (lead); original draft (lead); review and editing (equal). Andrielle Larissa Tchamba Kemajou: field activities (equal); methodology (equal); formal analysis (supporting); original draft (supporting); review and editing (supporting). Nancielle Mbiatong Tchatat: methodology (equal); field activities (equal); review and editing (supporting). Manuel Ritter: conceptualization (supporting), review and editing (equal). Bertrand Tatsinkou Fossi: conceptualization (lead), supervision (lead), review and editing (equal). Samuel Wanji: conceptualization (lead), supervision, review and editing (equal). All the authors have read and agreed to the final manuscript.
The authors are grateful to the head of Research Foundation for Tropical Diseases and the Environment (REFOTDE) for providing resources and also the staff of the Microbiology Unit at REFOTDE. We are grateful for the cooperation of the aquaculture facility owner and farm attendants during sample collection.
Table 1: distribution of isolated enterobacterial species
Figure 1: pooled total heterotrophic aerobic bacterial load by sample unit across ponds and collection time points Data represent the Log10 (± SD) CFU per gram or ml of sample. Data columns with different superscripts are significantly different from each other at p<0.05
Figure 2: antibiotic resistance profile of isolates (A) across ponds and (B) across collection time points (AMX (Amoxicillin), AM (Ampicillin), CRO (Ceftriaxone), CXM (Cefuroxime), IPM (Imipenem), SXT (Sulfamethoxazole and Trimethoprim), E (Erythromycin), CN (Gentamicin), TE (Tetracycline), NIT (Nitrofurantoin), OFX (Ofloxacin))
Figure 3: most frequent multiple drug resistant profile distributed across (A) fish parts and pond environment; (B) ponds and (C) collection time points (AMX (Amoxicillin), AM (Ampicillin), SXT (Sulfamethoxazole and Trimethoprim), CXM (Cefuroxime), CRO (Ceftriaxone), IPM (Imipenem), TE (Tetracycline), E (Erythromycin), NIT (Nitrofurantoin))
- Food and Agriculture Organization. Aquaculture. Accessed June 11, 2020.
- Santos L, Ramos F. Antimicrobial resistance in aquaculture: current knowledge and alternatives to tackle the problem. Int J Antimicrob Agents. 2018;52(2):135-143. PubMed | Google Scholar
- Food and Agriculture Organization. Is the planet approaching peak fish? Not so fast, study says. Accessed September 4, 2020.
- Mkong Cynthia Jeh, Molua Ernest Lytia, Mvodo Stephanie. Determinants of profitability of fish farming in Cameroon. Agriculture, Forestry and Fisheries. 2018;7(3):89. Google Scholar
- Pulkkinen K, Suomalainen LR, Read AF, Ebert D, Rintamäki P, Valtonen ET. Intensive fish farming and the evolution of pathogen virulence: the case of columnaris disease in Finland. Proc Biol Sci. 2010 Feb 22;277(1681):593-600. PubMed | Google Scholar
- Martins AFM, Pinheiro TL, Imperatori A, Freire SM, Sa-Freire L, Moreira BM et al. Plesiomonas shigelloides: a notable carrier of acquired antimicrobial resistance in small aquaculture farms. Aquaculture. 2019;500:514-520. Google Scholar
- Nicholson P, Fathi M, Fischer A, Mohan C, Schieck E, Mishra N et al. Detection of tilapia lake virus in Egyptian fish farms experiencing high mortalities in 2015. J Fish Dis. 2017;40(12):1925-1928. PubMed | Google Scholar
- Osman KM, Al-Maary KS, Aymen SM, Dawoud TM, Moussa IMI, Ibrahim MDS et al. Characterization and susceptibility of streptococci and enterococci isolated from Nile tilapia (oreochromis niloticus) showing septicaemia in aquaculture and wild sites in Egypt. BMC Vet Res. 2017 Nov 25;13(1):357. PubMed | Google Scholar
- Pakingking Jr R, Palma P, Usero R. Quantitative and qualitative analyses of the bacterial microbiota of tilapia (oreochromis niloticus) cultured in earthen ponds in the Philippines. World J Microbiol Biotechnol. 2015 Feb;31(2):265-275. PubMed | Google Scholar
- Kaktcham PM, Temgoua JB, Zambou FN, Diaz-Ruiz G, Wacher C, Pérez-Chabela ML. Quantitative analyses of the bacterial microbiota of rearing environment, tilapia and common carp cultured in earthen ponds and inhibitory activity of its lactic acid bacteria on fish spoilage and pathogenic bacteria. World J Microbiol Biotechnol. 2017 Feb;33(2):32. PubMed | Google Scholar
- Newaj-Fyzul A, Mutani A, Ramsubhag A, Adesiyun A. Prevalence of bacterial pathogens and their anti-microbial resistance in tilapia and their pond water in Trinidad. Zoonoses Public Health. 2008;55(4):206-213. PubMed | Google Scholar
- Economou V, Gousia P. Agriculture and food animals as a source antimicrobial-resistant bacteria. Infect Drug Resist. 2015 Apr 1;8:49-61. PubMed | Google Scholar
- Burridge L, Weis JS, Cabello F, Pizarro J, Bostick K. Chemical use in salmon aquaculture: a review of current practices and possible environmental effects. Aquaculture. 2010;306(1-4):7-23. Google Scholar
- Cabello FC, Godfrey HP, Buschmann AH, Dölz HJ. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect Dis. 2016 Jul;16(7):127-133. PubMed | Google Scholar
- Meek RW, Vyas H, Piddock LJV. Nonmedical uses of antibiotics: time to restrict their use? PLoS Biol. 2015;13(10):e1002266. PubMed | Google Scholar
- World Health Organization. Stop using antibiotics in healthy animals to preserve their effectiveness. Accessed March 1, 2020.
- Done HY, Venkatesan AK, Halden RU. Does the recent growth of aquaculture create antibiotic resistance threats different from those associated with land animal production in agriculture? AAPS J. 2015 May;17(3):513-524. PubMed | Google Scholar
- Manga VE, Agyingi CM, Djieto-Lordon AE. In-channel sand extraction in River Mungo, Cameroon: nature effects and concerns. ICWRER Proceedings. 2013;11-27. Google Scholar
- Tening AS, Chuyong GB, Asongwe GA, Fonge BA, Lifongo LL, Mvondo-Ze AD et al. Contribution of some water bodies and the role of soils in the physicochemical enrichment of the Douala-Edea mangrove ecosystem. Afr J Environ Sci Technol. 2013;7(5):336-349. Google Scholar
- Molua EL, Lambi CM. Climate, hydrology and water resources in Cameroon. Environmental issues: problems and prospects. 2001. Bamenda: Unique Printers. Google Scholar
- Boyd CE, Wood CW, Thunjai T. Aquaculture pond bottom soil quality management. 2002;1-45. Google Scholar
- Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493-496. PubMed | Google Scholar
- Patel JB, Cockerill FR, Bradford PA. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. Clinical and Laboratory Standard Institute. 2015;35(3):29-50. Google Scholar
- Al-Harbi Ah, Uddin N. Bacterial diversity of tilapia (oreochromis niloticus) cultured in brackish water in Saudi Arabia. Aquaculture. 2005;250(3-4):566-572. Google Scholar
- Shinkafi SA, Ukwaja VC. Bacteria associated with fresh tilapia fish (oreochromis niloticus) sold at Sokoto. Nigerian Journal of Basic and Applied Science. 2010;18(2):217-221. Google Scholar
- Rekhari YC, Agrawal R, Trakroo MD, Tiwari H. Qualitative and quantitative study on bacterial flora of farm raised common carp, Cyprinus carpio in India. Afr J Microbiol Res. 2014;8(11):1125-1129. Google Scholar
- Thanh Huong, Thuy Hien, Gallardo Wenresti, Thanh Hai. 2014 Bacterial population in intensive tilapia culture pond sediment in Hai. Int J Fish Aquac. 2014;6(12):133-139. PubMed | Google Scholar
- Daisy KR. The bacterial flora of tilapia (oleochromis niloticus) and catfish (clarias gariepinus) from earthen ponds in Sagana fish farm and Masinga Dam. Thesis. 2015.
- Markosova R, Jezek J. Indicator bacteria and limnological parameters in fish ponds. Wat Res. 1994;28(12):2477-2485. Google Scholar
- Donkeng NN, Maiwore J, Ngoune LT, Montet D, Mbofung CMF. Characterization of the bacterial flora of tilapia (Oreochoromis niloticus) harvested from four lakes in the north of Cameroon. African Journal of Biotechnology. 2011;10(71):16016-16023. Google Scholar
- Musefiu TA, Obuko EB, Bolarinwa A. Isolation and identification of aerobic bacteria flora of the skin and stomach of wild and cultured clarias gariepinus and oreochromis niloticus from Ibadan, Southwest Nigeria. Journal of Apllied Sciences Research. 2011;7(7)1047-1051.
- Jane-Francis TKA, Bughe RN, Oben BO, Ndip Lucy, Ndip LM, Ndip RN. Phenotypic characterization of human pathogenic bacteria in fish from the Coastal waters of South West Cameroon. Rev Environ Health. 2009;24(2):147-56. PubMed | Google Scholar
- Angulo FJ, Nargund VN, Chiller TC. Evidence of an association between use of anti-microbial agents in food animals and anti-microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. J Vet Med B Infect Dis Vet Public Health. Oct-Nov 2004;51(8-9):374-9. PubMed | Google Scholar
- Gauthier DT. Bacterial zoonoses of fishes: a review and appraisal of evidence for linkages between fish and human infections. Vet J. 2015;203(1):27-35. PubMed | Google Scholar
- Singer AC, Shaw H, Rhodes V, Hart A. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front Microbiol. 2016 Nov 1;7:1728. PubMed | Google Scholar
- Singh R, Singh AP, Kumar S, Giri BS, Kim KH. Antibiotic resistance in major rivers in the world: a systematic review on occurrence, emergence, and management strategies. J Clean Prod. 2019;234:1484-1505. Google Scholar
- Dib AL, Agabou A, Chahed Amina, Kurekci C, Moreno E, Espigares M, Elena E. Isolation, molecular characterization and antimicrobial resistance of enterobacteriaceae isolated from fish and seafood. Food Control. 2018;88:54-60. PubMed | Google Scholar
- World Health Organization. Critically important antimicrobials for human medicine. 5th revision 2016. Accessed March 1, 2020.
- Ntsama ISB, Tambe BA, Takadong JJT, Nama GM, Kansci G. Characteristics of fish farming practices and agrochemicals usage therein in four regions of Cameroon. Egypt J Aquat Res. 2018;44(2):145-153. Google Scholar
- Akinbowale OL, Peng H, Barton MD. Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. J Appl Microbiol. 2006;100(5):1103-1113. PubMed | Google Scholar
- Kathleen MM, Samuel L, Felecia C, Reagan EL, Kasing A, Lesley M, Toh SC. Antibiotic resistance of diverse bacteria from aquaculture in Borneo. Int J Microbiol. 2016;2016:2164761. PubMed | Google Scholar
- Lee SW, Wendy W. Antibiotic and heavy metal resistance of Aeromonas hydrophila and Edwardsiella tarda isolated from red hybrid tilapia (Oreochromis spp.) coinfected with motile aeromonas septicemia and edwardsiellosis. Vet World. 2017;10(7):803-807. PubMed | Google Scholar
- Wamala SP, Mugimba KK, Mutoloki S, Evensen ø, Mdegela R, Byarugaba DK, Sørum H. Occurrence and antibiotic susceptibility of fish bacteria isolated from oreochromis niloticus (Nile tilapia) and clarias gariepinus (African catfish) in Uganda. Fish Aquatic Sci. 2018;21(1):6. Google Scholar
- Watts JEM, Schreier HJ, Lanska L, Hale MS. The rising tide of antimicrobial resistance in aquaculture: sources, sinks and solutions. Mar Drugs. 2017;15(6):158. PubMed | Google Scholar
Search
This article authors
On Pubmed
On Google Scholar
Citation [Download]
Navigate this article
Similar articles in
Key words
Tables and figures
 Figure 2: antibiotic resistance profile of isolates (A) across ponds and (B) across collection time points. (AMX (Amoxicillin), AM (Ampicillin), CRO (Ceftriaxone), CXM (Cefuroxime), IPM (Imipenem), SXT (Sulfamethoxazole and Trimethoprim), E (Erythromycin), CN (Gentamicin), TE (Tetracycline), NIT (Nitrofurantoin), OFX (Ofloxacin))
Figure 2: antibiotic resistance profile of isolates (A) across ponds and (B) across collection time points. (AMX (Amoxicillin), AM (Ampicillin), CRO (Ceftriaxone), CXM (Cefuroxime), IPM (Imipenem), SXT (Sulfamethoxazole and Trimethoprim), E (Erythromycin), CN (Gentamicin), TE (Tetracycline), NIT (Nitrofurantoin), OFX (Ofloxacin))
 Figure 3: most frequent multiple drug resistant profile distributed across (A) fish parts and pond environment; (B) ponds and (C) collection time points (AMX (Amoxicillin), AM (Ampicillin), SXT (Sulfamethoxazole and Trimethoprim), CXM (Cefuroxime), CRO (Ceftriaxone), IPM (Imipenem), TE (Tetracycline), E (Erythromycin), NIT (Nitrofurantoin))
Figure 3: most frequent multiple drug resistant profile distributed across (A) fish parts and pond environment; (B) ponds and (C) collection time points (AMX (Amoxicillin), AM (Ampicillin), SXT (Sulfamethoxazole and Trimethoprim), CXM (Cefuroxime), CRO (Ceftriaxone), IPM (Imipenem), TE (Tetracycline), E (Erythromycin), NIT (Nitrofurantoin))